Page 82 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 82
54 Part I Molecular and Cellular Basis of Hematology
concomitant release of the GPI signal peptide. The GPI attachment the CNX/CRT chaperone system. In a number of cases retrotranslo-
promotes membrane association. cation appears to require reduction of disulfide bridges by PDI.
Similarly, BiP association with substrates (e.g., unassembled immu-
Destruction of Misfolded or Misassembled noglobulin light chains) can direct them to ERAD. If a protein
Proteins: Endoplasmic-Reticulum Associated remains in its unfolded state for an extended period of time, trim-
ming of the Man 8GlcNac 2 also occurs. This processing is catalyzed
Degradation (ERAD) by ER-degradation enhancer mannosidase α-like proteins EDEM1,
EDEM2, EDEM3 (Htm1p in yeast). The current model postulates
In the ER, proteins undergo a so-called quality control, which ensures that N-glycan structure generated by extensive demannosylation is
that only correctly folded proteins exit the ER. Consequently, mis- the signal for glycoprotein degradation. ER-resident lectins (OS-9
folded proteins are extracted from the ER folding environment for and XTP3-B) bind to the remaining mannose residues and assist the
disposal. This mode of degradation is referred to as endoplasmic retrotranslocation.
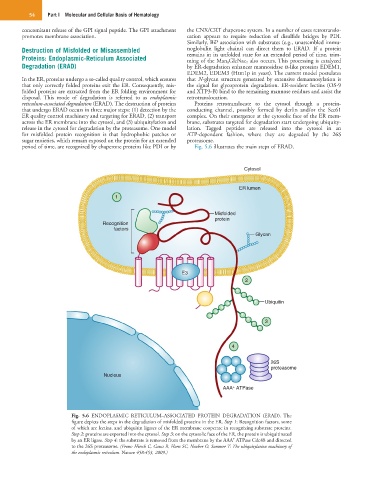
reticulum-associated degradation (ERAD). The destruction of proteins Proteins retrotranslocate to the cytosol through a protein-
that undergo ERAD occurs in three major steps: (1) detection by the conducting channel, possibly formed by derlin and/or the Sec61
ER quality control machinery and targeting for ERAD, (2) transport complex. On their emergence at the cytosolic face of the ER mem-
across the ER membrane into the cytosol, and (3) ubiquitylation and brane, substrates targeted for degradation start undergoing ubiquity-
release in the cytosol for degradation by the proteasome. One model lation. Tagged peptides are released into the cytosol in an
for misfolded protein recognition is that hydrophobic patches or ATP-dependent fashion, where they are degraded by the 26S
sugar moieties, which remain exposed on the protein for an extended proteasome.
period of time, are recognized by chaperone proteins like PDI or by Fig. 5.6 illustrates the main steps of ERAD.
Cytosol
ER lumen
1
Misfolded
protein
Recognition
factors
Glycan
E3
2
Ubiquitin
3
4
26S
proteasome
Nucleus
+
AAA ATPase
Fig. 5.6 ENDOPLASMIC RETICULUM–ASSOCIATED PROTEIN DEGRADATION (ERAD). The
figure depicts the steps in the degradation of misfolded proteins in the ER. Step 1: Recognition factors, some
of which are lectins, and ubiquitin ligases of the ER membrane cooperate in recognizing substrate proteins.
Step 2: proteins are exported into the cytosol. Step 3: on the cytosolic face of the ER, the protein is ubiquitinated
+
by an ER ligase. Step 4: the substrate is removed from the membrane by the AAA ATPase Cdc48 and directed
to the 26S proteasome. (From: Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T: The ubiquitylation machinery of
the endoplasmic reticulum. Nature 458:453, 2009.)