Page 984 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 984
Chapter 57 Pharmacology and Molecular Mechanisms of Antineoplastic Agents for Hematologic Malignancies 867
the degradation of intracellular proteins. Numerous studies have least three distinct proteolytic activities are associated with the protea-
demonstrated that the ubiquitin–proteasome system controls basic some: chymotryptic, tryptic, and peptidylglutamyl. After release from
cellular functions such as cell cycle progression, signal transduc- the substrate, the polyubiquitin chain is hydrolyzed into single
tion, and programmed cell death, hence the interest in therapeutic ubiquitin moieties, and tagged proteins are degraded to small pep-
interventions that manipulate proteasomal activity and potentially tides. Both the assembly of the 26S proteasome and the degradation
restore cellular homeostasis into transformed cells (see Chapter 4). of protein substrates are ATP dependent. The mechanism leading to
Ubiquitin is a highly conserved 76-amino–acid polypeptide that cell death after proteasome inhibition are not fully understood, but
is expressed in all eukaryotic cells. Under the sequential action of E1 it is hypothesized that accumulation of incompatible regulatory
(ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme), proteins within the cell, accumulation of mis-folded proteins from
and E3 (ubiquitin ligase), ubiquitin is activated and covalently con- the ER (“ER stress”) leading to suspension of protein synthesis, and
jugated to potential proteasome substrates via an isopeptide bond interference with degradation of proteins that inhibit NFκB, leading
between the C-terminal glycine residue of ubiquitin and the ε-amino to inhibition of NFκB, are the major contributors to cell death
group of internal lysine residues in target proteins. The same set of induced by proteasome inhibitors. Synergism between proteasome
enzymes also catalyzes the formation of the isopeptide bond between inhibition and cytotoxic chemotherapy is an area of active research.
G76 and the lysine residue (K48) of previously conjugated ubiquitin, The activated B-cell subtype of DLBCL (ABC-DLBCL), which has
leading to formation of a polyubiquitin chain. Polyubiquitinated inferior survival after anthracycline-based chemotherapy, is character-
substrates are usually targeted for proteasomal degradation. ized by constitutive activation of the NFκB pathway, leading to
The 26S proteasome is a large (2000-kDa) threonine protease resistance to apoptosis via the mitochondrial pathway through
present in the nucleus and cytoplasm of all eukaryotic cells. This upregulation of NFκB-regulated genes (including bcl-2, bcl-XL,
ATP-dependent, multicatalytic protease eliminates damaged or mis- X-linked inhibitor of apoptosis [XIAP], and survivin). This explains
folded proteins and regulates cyclins and CDK inhibitor cell cycle the clinical benefit seen with the addition of bortezomib, the first-in-
regulatory proteins as well as other proteins that govern the transcrip- class proteasome inhibitor to anthracycline-based chemotherapy in
tion factor activation, apoptosis, and cell trafficking. Its structure ABC-DLBCL.
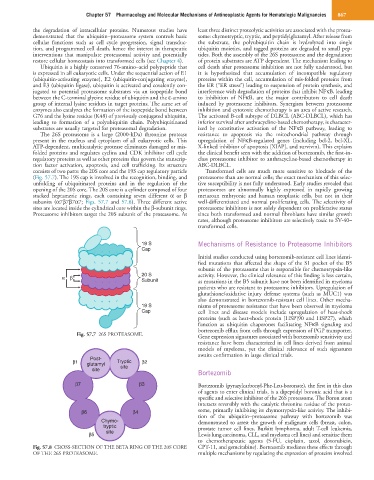
consists of two parts: the 20S core and the 19S cap regulatory particle Transformed cells are much more sensitive to blockade of the
(Fig. 57.7). The 19S cap is involved in the recognition, binding, and proteasome than are normal cells; the exact mechanism of this selec-
unfolding of ubiquitinated proteins and in the regulation of the tive susceptibility is not fully understood. Early studies revealed that
opening of the 20S core. The 20S core is a cylinder composed of four proteasomes are abnormally highly expressed in rapidly growing
stacked heptameric rings, each containing seven different α or β metazoan embryonic and human neoplastic cells, but not in their
subunits (α7β7β7α7; Figs. 57.7 and 57.8). Three different active well-differentiated and normal proliferating cells. The selectivity of
sites are located inside the cylindrical core within the β-subunit rings. proteasome inhibitors is not solely dependent on proliferative status
Proteasome inhibitors target the 20S subunit of the proteasome. At since both transformed and normal fibroblasts have similar growth
rates, although proteasome inhibitors are selectively toxic to SV-40–
transformed cells.
19 S Mechanisms of Resistance to Proteasome Inhibitors
Cap
Initial studies conducted using bortezomib-resistant cell lines identi-
fied mutations that affected the shape of the S1 pocket of the B5
subunit of the proteasome that is responsible for chemotrypsin-like
20 S activity. However, the clinical relevance of this finding is less certain,
α β Subunit as mutations in the B5 subunit have not been identified in myeloma
patients who are resistant to proteasome inhibitors. Upregulation of
glutathione/oxidative injury defense systems (such as MUC1) was
also demonstrated in bortezomib-resistant cell lines. Other mecha-
19 S nisms of proteasome resistance that have been observed in myeloma
Cap cell lines and disease models include upregulation of heat-shock
proteins (such as heat-shock protein [HSP]90 and HSP27), which
function as ubiquitin chaperones facilitating NFκB signaling and
bortezomib efflux from cells through expression of PGP transporter.
Fig. 57.7 26S PROTEASOME. Gene expression signatures associated with bortezomib sensitivity and
resistance have been characterized in cell lines derived from animal
models of myeloma, yet the clinical relevance of such signatures
awaits confirmation in large clinical trials.
Post-
β1 glutamyl Tryptic β2
site site
Bortezomib
β7 β3 Bortezomib (pyrazylcarbonyl-Phe-Leu-boronate), the first in this class
of agents to enter clinical trials, is a dipeptidyl boronic acid that is a
specific and selective inhibitor of the 26S proteasome. The Boron atom
interacts reversibly with the catalytic threonine residue of the protea-
β6 β4 some, primarily inhibiting its chymotrypsin-like activity. The inhibi-
tion of the ubiquitin–proteasome pathway with bortezomib was
Chymo- demonstrated to arrest the growth of malignant cells (breast, colon,
tryptic prostate tumor cell lines, Burkitt lymphoma, adult T-cell leukemia,
site
β5 Lewis lung carcinoma, CLL, and myeloma cell lines) and sensitize them
to chemotherapeutic agents (5-FU, cisplatin, taxol, doxorubicin,
Fig. 57.8 CROSS-SECTION OF THE BETA RING OF THE 20S CORE CPT-11, and gemcitabine). Bortezomib mediates these effects through
OF THE 26S PROTEASOME. multiple mechanisms by regulating the expression of proteins involved