Page 1830 - Williams Hematology ( PDFDrive )
P. 1830
1804 Part XI: Malignant Lymphoid Diseases Chapter 110: Heavy-Chain Disease 1805
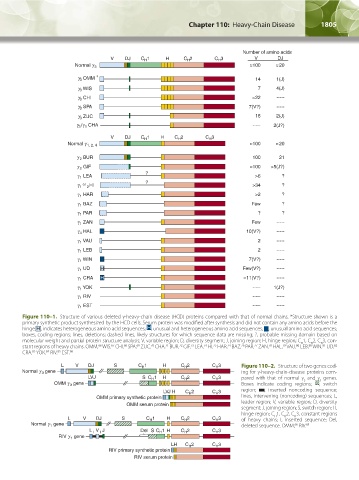
Figure 110–1. Structure of various deleted γ-heavy-chain disease (HCD) proteins compared with that of normal chains. *Structure shown is a
primary synthetic product synthesized by the HCD cells. Serum protein was modified after synthesis and did not contain any amino acids before the
hinge. H , indicates heterogeneous amino acid sequences; H , unusual and heterogeneous amino acid sequences; , unusual amino acid sequences;
boxes, coding regions; lines, deletions; dashed lines, likely structures for which sequence data are missing; ?, probable missing domain based on
molecular weight and partial protein structure analysis; V, variable region; D, diversity segment; J, joining region; H, hinge region; C 1, C 2, C 3, con-
H
H
H
stant regions of heavy chains. OMM, WIS, CHI, SPA, ZUC, CHA, BUR, GIF, LEA, HI, HAR, BAZ, PAR, ZAN, HAL, VAU, LEB, WIN, UD,
81
74
74
76
75
78
77
79
80
80
68
69
67
82
66
71
72
73
70
CRA, YOK, RIV, EST. 86
83
85
84
Figure 110–2. Structure of two genes cod-
ing for γ-heavy-chain-disease proteins com-
pared with that of normal γ and γ genes.
3
1
Boxes indicate coding regions; , switch
region; , inserted noncoding sequence;
lines, intervening (noncoding) sequences; L,
leader region; V, variable region; D, diversity
segment; J, joining region; S, switch region; H,
hinge region; C 1, C 2, C 3, constant regions
H
H
H
of heavy chains; I, inserted sequence; Del,
deleted sequence. OMM, RIV. 85
66
Kaushansky_chapter 110_p1803-1812.indd 1805 9/18/15 9:57 AM