Page 2332 - Williams Hematology ( PDFDrive )
P. 2332
2306 Part XII: Hemostasis and Thrombosis Chapter 135: Fibrinolysis and Thrombolysis 2307
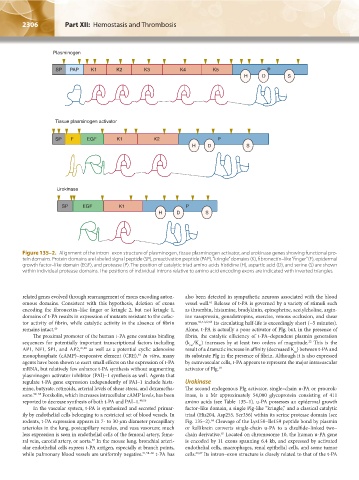
Plasminogen
SP PAP K1 K2 K3 K4 K5 P
H D S
Tissue plasminogen activator
SP F EGF K1 K2 P
H D S
Urokinase
SP EGF K1 P
H D S
Figure 135–2. Alignment of the intron–exon structure of plasminogen, tissue plasminogen activator, and urokinase genes showing functional pro-
tein domains. Protein domains are labeled signal peptide (SP), preactivation peptide (PAP), “kringle” domains (K), fibronectin–like “finger” (F), epidermal
growth factor–like domain (EGF), and protease (P). The position of catalytic triad amino acids histidine (H), aspartic acid (D), and serine (S) are shown
within individual protease domains. The positions of individual introns relative to amino acid encoding exons are indicated with inverted triangles.
related genes evolved through rearrangement of exons encoding auton- also been detected in sympathetic neurons associated with the blood
omous domains. Consistent with this hypothesis, deletion of exons vessel wall. Release of t-PA is governed by a variety of stimuli such
61
encoding the fibronectin–like finger or kringle 2, but not kringle 1, as thrombin, histamine, bradykinin, epinephrine, acetylcholine, argin-
domains of t-PA results in expression of mutants resistant to the cofac- ine vasopressin, gonadotropins, exercise, venous occlusion, and shear
tor activity of fibrin, while catalytic activity in the absence of fibrin stress. 50,51,62,63 Its circulating half-life is exceedingly short (~5 minutes).
remains intact. 46 Alone, t-PA is actually a poor activator of Plg, but, in the presence of
The proximal promoter of the human t-PA gene contains binding fibrin, the catalytic efficiency of t-PA–dependent plasmin generation
sequences for potentially important transcriptional factors including (k /K ) increases by at least two orders of magnitude. This is the
22
m
cat
AP1, NF1, SP1, and AP2, 47,48 as well as a potential cyclic adenosine result of a dramatic increase in affinity (decreased K ) between t-PA and
m
monophosphate (cAMP)–responsive element (CRE). In vitro, many its substrate Plg in the presence of fibrin. Although it is also expressed
49
agents have been shown to exert small effects on the expression of t-PA by extravascular cells, t-PA appears to represent the major intravascular
mRNA, but relatively few enhance t-PA synthesis without augmenting activator of Plg. 18
plasminogen activator inhibitor (PAI)–1 synthesis as well. Agents that
regulate t-PA gene expression independently of PAI–1 include hista- Urokinase
mine, butyrate, retinoids, arterial levels of shear stress, and dexametha- The second endogenous Plg activator, single–chain u-PA or prourok-
sone. 50–55 Forskolin, which increases intracellular cAMP levels, has been inase, is a Mr approximately 54,000 glycoprotein consisting of 411
reported to decrease synthesis of both t-PA and PAI–1. 48,56 amino acids (see Table 135–1). u-PA possesses an epidermal growth
In the vascular system, t-PA is synthesized and secreted primar- factor–like domain, a single Plg-like “kringle,” and a classical catalytic
ily by endothelial cells belonging to a restricted set of blood vessels. In triad (His204, Asp255, Ser356) within its serine protease domain (see
rodents, t-PA expression appears in 7- to 30-μm diameter precapillary Fig. 135–2). Cleavage of the Lys158–Ile159 peptide bond by plasmin
64
arterioles in the lung, postcapillary venules, and vasa vasorum; much or kallikrein converts single-chain u-PA to a disulfide–linked two–
less expression is seen in endothelial cells of the femoral artery, femo- chain derivative. Located on chromosome 10, the human u-PA gene
65
ral vein, carotid artery, or aorta. In the mouse lung, bronchial arteri- is encoded by 11 exons spanning 6.4 kb, and expressed by activated
57
olar endothelial cells express t-PA antigen, especially at branch points, endothelial cells, macrophages, renal epithelial cells, and some tumor
while pulmonary blood vessels are uniformly negative. 51,58–60 t-PA has cells. 66,67 Its intron–exon structure is closely related to that of the t-PA
Kaushansky_chapter 135_p2303-2326.indd 2306 9/18/15 5:13 PM