Page 1153 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 1153
1012 Part VII Hematologic Malignancies
The oncogenic activity of MLL fusions also requires their physical
interaction with Menin, the tumor suppressor protein encoded by
DOT1L Complex AF10 DOT1L ENL N-terminal domain of MLL that is conserved in all MLL fusion
the multiple endocrine neoplasia 1 (MEN1) gene. Menin binds an
AF17
275,276
Moreover, abrogating the interaction of Menin with
proteins.
AF9
MLL, either by deleting Menin or by mutating the Menin-binding
domain of MLL, impairs both transcriptional and oncogenic func-
275,277
tions of MLL fusions.
These findings spurred the development
of small-molecule inhibitors of the MLL–Menin interaction, and one
ENL/AF9 AF4/AF5q31 such inhibitor has demonstrated promising activity in preclinical
278
models, both in vitro and in vivo.
ELL1-3 The presence of MLL rearrangements is associated with dismal
SEC P – T E F b EAF outcomes despite aggressive chemotherapy in most cases of
B-precursor ALL. 16,279–281 Although bone marrow transplantation
(BMT) in first remission does not appear to improve outcomes for
all infants with MLL-rearranged ALL, 281,282 a recent comprehensive
study revealed significantly improved outcomes with BMT in infants
AF4/AF5q31
BRD4 Complex P – T E F b EAF Recent data demonstrating the therapeutic utility of small-molecule
283
with MLL-rearranged ALL who have particularly high-risk features.
ELL1-3
inhibitors of DOT1L and of the MLL–Menin interaction have gener-
ated considerable enthusiasm for the application of these novel thera-
peutic strategies for patients with MLL-rearranged leukemias.
BRD4 PAFc
PRC2 Mutations in T-Cell ALL
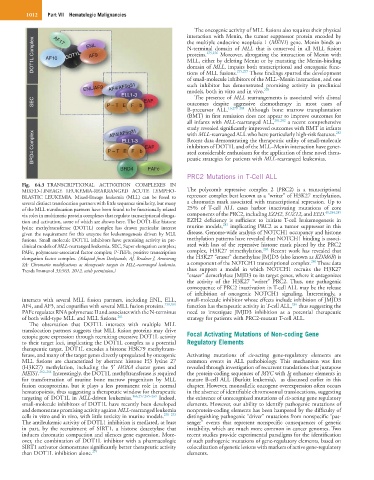
Fig. 64.3 TRANSCRIPTIONAL ACTIVATION COMPLEXES IN
MIXED-LINEAGE LEUKEMIA-REARRANGED ACUTE LYMPHO- The polycomb repressive complex 2 (PRC2) is a transcriptional
BLASTIC LEUKEMIA. Mixed-lineage leukemia (MLL) can be fused to repressor complex best known as a “writer” of H3K27 methylation,
several distinct translocation partners with little sequence similarity, but many a chromatin mark associated with transcriptional repression. Up to
of the MLL translocation partners have been found to be functionally related 25% of T-cell ALL cases harbor inactivating mutations of core
40,284,285
via roles in multimeric protein complexes that regulate transcriptional elonga- components of the PRC2, including EZH2, SUZ12, and EED.
tion and activation, some of which are shown here. The DOT1-like histone EZH2 deficiency is sufficient to initiate T-cell leukemogenesis in
285
lysine methyltransferase (DOT1L) complex has drawn particular interest murine models, implicating PRC2 as a tumor suppressor in this
given the requirement for this enzyme for leukemogenesis driven by MLL disease. Genome-wide analysis of NOTCH1 occupancy and histone
fusions. Small molecule DOT1L inhibitors have promising activity in pre- methylation patterns have revealed that NOTCH1 binding is associ-
clinical models of MLL-rearranged leukemia. SEC, Super elongation complex; ated with loss of the repressive histone mark placed by the PRC2
284
PAFc, polymerase-associated factor complex; P-TEFb, positive transcription complex, H3K27 trimethylation. Recent work has revealed that
elongation factor complex. (Adapted from Deshpande, AJ, Bradner J, Armstrong the H3K27 “eraser” demethylase JMJD3 (also known as KDM6B) is
286
SA: Chromatin modifications as therapeutic targets in MLL-rearranged leukemia. a component of the NOTCH1 transcriptional complex. These data
Trends Immunol 33:563, 2012, with permission.) thus support a model in which NOTCH1 recruits the H3K27
“eraser” demethylase JMJD3 to its target genes, where it antagonizes
the activity of the H3K27 “writer” PRC2. Thus, one pathogenic
consequence of PRC2 inactivation in T-cell ALL may be the release
of inhibition of oncogenic NOTCH1 signaling. Interestingly, a
interacts with several MLL fusion partners, including ENL, ELL, small-molecule inhibitor whose effects include inhibition of JMJD3
286
AF4, and AF5, and copurifies with several MLL fusion proteins. 259,260 function has therapeutic activity in T-cell ALL, thus suggesting the
PAFc regulates RNA polymerase II and associates with the N-terminus need to investigate JMJD3 inhibition as a potential therapeutic
of both wild-type MLL and MLL fusions. 261 strategy for patients with PRC2-mutant T-cell ALL.
The observation that DOT1L interacts with multiple MLL
translocation partners suggests that MLL fusion proteins may drive
ectopic gene expression through recruiting excessive DOT1L activity Focal Activating Mutations of Non-coding Gene
to their target loci, implicating the DOT1L complex as a potential Regulatory Elements
therapeutic target. DOT1L encodes a histone H3K79 methyltrans-
ferase, and many of the target genes directly upregulated by oncogenic Activating mutations of cis-acting gene-regulatory elements are
MLL fusions are characterized by aberrant histone H3 lysine 27 common events in ALL pathobiology. This mechanism was first
(H3K27) methylation, including the 5′ HOXA cluster genes and revealed through investigation of recurrent translations that juxtapose
MEIS1. 262–264 Interestingly, the DOT1L methyltransferase is required the protein-coding sequences of MYC with Ig enhancer elements in
for transformation of murine bone marrow progenitors by MLL mature B-cell ALL (Burkitt leukemia), as discussed earlier in this
fusion oncoproteins, but it plays a less prominent role in normal chapter. However, monoallelic oncogene overexpression often occurs
hematopoiesis, thus suggesting a therapeutic window for therapeutic in the absence of identifiable chromosomal translocations, suggesting
targeting of DOT1L in MLL-driven leukemias. 166,257,265–269 Indeed, the existence of unrecognized mutations of cis-acting gene regulatory
small-molecule inhibitors of DOT1L have recently been developed elements. However, our ability to identify pathogenic mutations of
and demonstrate promising activity against MLL-rearranged leukemia nonprotein-coding elements has been hampered by the difficulty of
cells in vitro and in vivo, with little toxicity in murine models. 270–273 distinguishing pathogenic “driver” mutations from nonspecific “pas-
The antileukemic activity of DOTL1 inhibition is mediated, at least senger” events that represent nonspecific consequences of genetic
in part, by the recruitment of SIRT1, a histone deacetylase that instability, which are much more common in cancer genomes. Two
induces chromatin compaction and silences gene expression. More- recent studies provide experimental paradigms for the identification
over, the combination of DOT1L inhibitor with a pharmacologic of such pathogenic mutations of gene-regulatory elements, based on
SIRT1 activator demonstrates significantly better therapeutic activity colocalization of genetic lesions with markers of active gene-regulatory
than DOT1L inhibition alone. 274 elements.