Page 32 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 32
4 Part I Molecular and Cellular Basis of Hematology
A B C
3′ end
3′ C:G 5′
5′ end H O A:T
O H 2′ 3′ H
5′ H C O N H G:C 5′ 3′
A:T
2
4′
C:G
4′ H H 1′ N H N N 1′ H O 5′CH T:A T A
H 3′ 2′ H O H N N 2 G:C A T
T:A
O H CH 3 O C:G G C
Thymine H N - A:T
- O P O Adenine O P O C G
A:T
O N H CH 3 H O C:G 3′ 5′
G:C
N H O H 2′ 3′ H T:A
5′ H C O N 1′ H H 4′
2
4′ H H 1′ N H N G T:A C 5′
N N O 5′ CH 2 3′ G C
H 3′ 2′ H Adenine O Thymine O C:G G
O H A:U T
T:A
- O P O O P O - C:G A
G
N H H O A:U T
O A:U T
O H N H 2′ 3′ H T:A A
5′ H C O N 1′ H H 4′ C:G G
2
4′ H H 1′ N H N G:C C
N N O 5′ CH 2 G:C A C
T
H 3′ 2′ H N H O O A A
T
O H Guanine H Cytosine A T
- O P O O P O - T A
H H O 5′ A T 3′
O H 2′ 3′ H
O H N A:T
5′ H C O N N 1′ H H 4′ G:C
2
C:G
4′ H H 1′ N H N O T:A
N 5′ CH 2
H 3′ 2′ H N H O G:C
T:A
O H N 5′ end C:G
- O P O Cytosine Guanine 5′ A:T 3′
3′ end
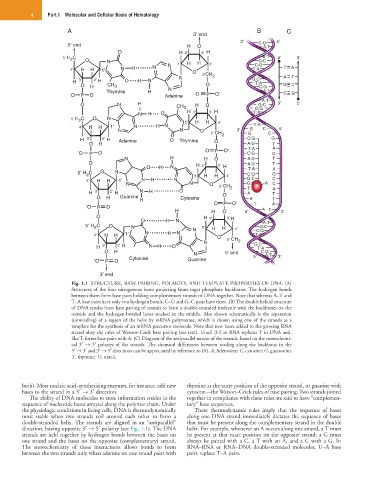
Fig. 1.1 STRUCTURE, BASE PAIRING, POLARITY, AND TEMPLATE PROPERTIES OF DNA. (A)
Structures of the four nitrogenous bases projecting from sugar phosphate backbones. The hydrogen bonds
between them form base pairs holding complementary strands of DNA together. Note that whereas A–T and
T–A base pairs have only two hydrogen bonds, C–G and G–C pairs have three. (B) The double helical structure
of DNA results from base pairing of strands to form a double-stranded molecule with the backbones on the
outside and the hydrogen-bonded bases stacked in the middle. Also shown schematically is the separation
(unwinding) of a region of the helix by mRNA polymerase, which is shown using one of the strands as a
template for the synthesis of an mRNA precursor molecule. Note that new bases added to the growing RNA
strand obey the rules of Watson–Crick base pairing (see text). Uracil (U) in RNA replaces T in DNA and,
like T, forms base pairs with A. (C) Diagram of the antiparallel nature of the strands, based on the stereochemi-
cal 3′ → 5′ polarity of the strands. The chemical differences between reading along the backbone in the
5′ → 3′ and 3′ → 5′ directions can be appreciated by reference to (A). A, Adenosine; C, cytosine; G, guanosine;
T, thymine; U, uracil.
both). Most nucleic acid–synthesizing enzymes, for instance, add new thymine at the same position of the opposite strand, or guanine with
bases to the strand in a 5′ → 3′ direction. cytosine—the Watson–Crick rules of base pairing. Two strands joined
The ability of DNA molecules to store information resides in the together in compliance with these rules are said to have “complemen-
sequence of nucleotide bases arrayed along the polymer chain. Under tary” base sequences.
the physiologic conditions in living cells, DNA is thermodynamically These thermodynamic rules imply that the sequence of bases
most stable when two strands coil around each other to form a along one DNA strand immediately dictates the sequence of bases
double-stranded helix. The strands are aligned in an “antiparallel” that must be present along the complementary strand in the double
direction, having opposite 3′ → 5′ polarity (see Fig. 1.1). The DNA helix. For example, whenever an A occurs along one strand, a T must
strands are held together by hydrogen bonds between the bases on be present at that exact position on the opposite strand; a G must
one strand and the bases on the opposite (complementary) strand. always be paired with a C, a T with an A, and a C with a G. In
The stereochemistry of these interactions allows bonds to form RNA–RNA or RNA–DNA double-stranded molecules, U–A base
between the two strands only when adenine on one strand pairs with pairs replace T–A pairs.