Page 47 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 47
Chapter 2 Epigenetics and Epigenomics 19
structure is the nucleosome, consisting of 147 bp of DNA double citrullination, ubiquitination, and ADP-ribosylation) that alter their
helix wrapped around the core histone octamer (Fig. 2.1C). Linker interactions with DNA and nuclear proteins (Fig. 2.2). Histone-
histones, primarily H1, bind the nucleosome at the entry and exit modifying enzymes are broadly classified as “writers,” such as histone
sites of the DNA and allow the formation of higher-order structure. methyltransferases (HMTs) and histone acetyltransferases (HATs)
Histone N-terminal domains are rich in lysine and arginine residues that add functional groups, or “erasers,” such as histone demethylases
that are subject to a variety of posttranslational modifications (see (HDMs) and histone deacetylases (HDACs). DNA-binding proteins
later). contain a variety of “reader” protein domains (including bromodo-
In addition to these major histones, dozens of minor histone mains, chromodomains, tudor domains, SANT domains) that have
variants have been identified and are highly evolutionarily conserved. increased affinity for modified histones. In this way, covalently modi-
Some minor variants have very specific roles in chromatin regulation. fied histones constitute a “histone code” that is a defining feature of
For example, histone H3–like centromere protein A (CENPA) is the dynamic epigenome. Each of the eight histones in a nucleosome
associated with centromeres. H2A.Z is associated with the promoters can harbor multiple covalent modifications, giving the histone code
and enhancers of actively transcribed genes. Histone H3.3 is associ- tremendous combinatorial complexity.
ated with the body of actively transcribed genes. Phosphorylated Trimethylation of H3 lysine 4 (H3K4me3) and trimethylation of
H2A.X is found in regions around double-stranded DNA breaks and H3 lysine 36 (H3K36me3) are both associated with transcriptional
recruits DNA repair machinery. activation. H3K4me3 occurs at the promoter of active genes, and
the degree of trimethylation is broadly correlated with transcrip-
tional activity of the gene. H3K36me3 is deposited by the lysine
COVALENT HISTONE MODIFICATIONS methyltransferase KMT2A (also known as MLL1) component of the
Mediator complex and occurs in the body of active genes. H3K36me3
Histones undergo a variety of posttranslational modifications (in- associates with elongating RNA polymerase II, thus marking
cluding methylation, acetylation, phosphorylation, SUMOylation, actively transcribed genes. Mono- and dimethylation of H3 lysine
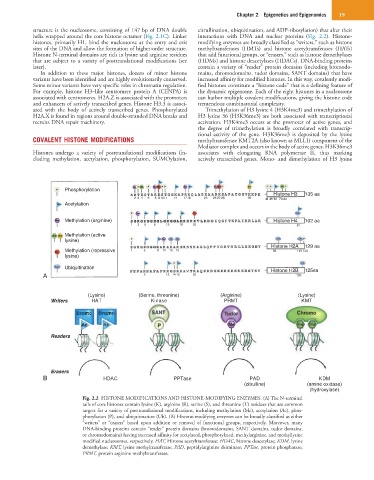
Phosphorylation
Histone H3 135 aa
2 3 4 6 8 9 10 11 14 1718 23 26 27 28 36 41 45 56 79 80
Acetylation
Methylation (arginine) Histone H4 102 aa
1 3 5 8 12 16 20 91
Methylation (active
lysine)
Histone H2A 129 aa
Methylation (repressive 1 5 9 11 13 15 63 119 120
lysine)
Ubiquitination Histone H2B 125aa
A 5 12 14 15 20 120
(Lysine) (Serine, threonine) (Arginine) (Lysine)
Writers HAT Kinase PRMT KMT
Readers
Erasers
B HDAC PPTase PAD KDM
(citrulline) (amine oxidase)
(hydroxylase)
Fig. 2.2 HISTONE MODIFICATIONS AND HISTONE-MODIFYING ENZYMES. (A) The N-terminal
tails of core histones contain lysine (K), arginine (R), serine (S), and threonine (T) residues that are common
targets for a variety of posttranslational modifications, including methylation (Me), acetylation (Ac), phos-
phorylation (P), and ubiquitination (Ub). (B) Histone-modifying enzymes can be broadly classified as either
“writers” or “erasers” based upon addition or removal of functional groups, respectively. Moreover, many
DNA-binding proteins contain “reader” protein domains (bromodomains, SANT domains, tudor domains,
or chromodomains) having increased affinity for acetylated, phosphorylated, methylarginine, and methyllysine
modified nucleosomes, respectively. HAT, Histone acetyltransferase; HDAC, histone deacetylase; KDM, lysine
demethylase; KMT, lysine methyltransferase; PAD, peptidylarginine deiminase; PPTase, protein phosphatase;
PRMT, protein arginine methyltransferase.