Page 46 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 46
18 Part I Molecular and Cellular Basis of Hematology
H3K4me1
H3K27ac H3K4me3 H3K36me3 CTCF H3K27me3 H3K9me3
p300
Enhancer Promoter Gene Insulator Gene cluster Repeats
Euchromatin Facultative Constitutive
A heterochromatin heterochromatin
Nucleosomes
Length: 2 m DNA 11 nm
Histone modifications
Histone H1
30 nm
Domain organization
C
300-700 nm
Enhancer
Mitotic condensation eRNA
Cohesin
mRNA
Length: <10 µm 1.5 µm
D Gene promoter
B Chromosome
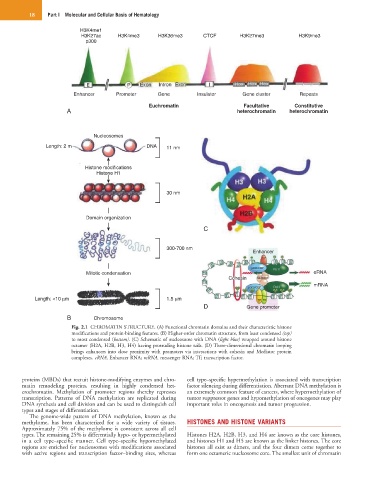
Fig. 2.1 CHROMATIN STRUCTURE. (A) Functional chromatin domains and their characteristic histone
modifications and protein-binding features. (B) Higher-order chromatin structure, from least condensed (top)
to most condensed (bottom). (C) Schematic of nucleosome with DNA (light blue) wrapped around histone
octamer (H2A, H2B, H3, H4) having protruding histone tails. (D) Three-dimensional chromatin looping
brings enhancers into close proximity with promoters via interactions with cohesin and Mediator protein
complexes. eRNA, Enhancer RNA; mRNA, messenger RNA; TF, transcription factor.
proteins (MBDs) that recruit histone-modifying enzymes and chro- cell type–specific hypermethylation is associated with transcription
matin remodeling proteins, resulting in highly condensed het- factor silencing during differentiation. Aberrant DNA methylation is
erochromatin. Methylation of promoter regions thereby represses an extremely common feature of cancers, where hypermethylation of
transcription. Patterns of DNA methylation are replicated during tumor suppressor genes and hypomethylation of oncogenes may play
DNA synthesis and cell division and can be used to distinguish cell important roles in oncogenesis and tumor progression.
types and stages of differentiation.
The genome-wide pattern of DNA methylation, known as the
methylome, has been characterized for a wide variety of tissues. HISTONES AND HISTONE VARIANTS
Approximately 75% of the methylome is consistent across all cell
types. The remaining 25% is differentially hypo- or hypermethylated Histones H2A, H2B, H3, and H4 are known as the core histones,
in a cell type–specific manner. Cell type–specific hypomethylated and histones H1 and H5 are known as the linker histones. The core
regions are enriched for nucleosomes with modifications associated histones all exist as dimers, and the four dimers come together to
with active regions and transcription factor–binding sites, whereas form one octameric nucleosome core. The smallest unit of chromatin