Page 535 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 535
450 Part V Red Blood Cells
ALA PRO Tyr HC2
LYS
C-Terminus ASN ALA GLN ALA PRO
HIS ALA ALA GLY GLN THR
LYS ALA VAL VAL TYR VAL PHE FG2 Val E11
HIS TYR VAL
LEU F9
140 H 130 GLU CD2
120 LYS
C7
G ALA GLY C3 M P CDI
ASN CYS HIS F8
VAL PHE H16 G1 C5
GLU ARG LEU VAL VAL LEU HIS CD7
100 GLY V
PROASN LEU LEU E7
PHE M
ASP 110 F1 C1 E1
G5 D1
VAL HIS D7
VAL N – Terminus LEU LYS ASP V
F 90 M
ALA
HIS GLU CYS E5
THR SER
GLY THR PHE LEU HIS
LEU LYS
LEU
THR LEU Proximal to Heme
GLU PRO 80 ASN HEME EF3 EF1 G15 B5
GLU LYS ASP NA1
SER E +
ALA A LEU HIS GLY 70 NH 3 E20 A16 B1
VAL ALA ASP Distal to Heme
THR ALA NA2
ALA LEU PHE H5 G19
LEU TRP SER GLY LYS AB1
GLY VAL HIS
LYS LEU LYS
VAL 20 Close Spatial Contact GLY ALA LYS A1
VALASP GLY LYS VAL H1
LEU LEU PRO
ASN VAL GLY ALA LEU 60 GH4
GLU GLY VAL ASN
GLU ARG TYR TRP C GLY
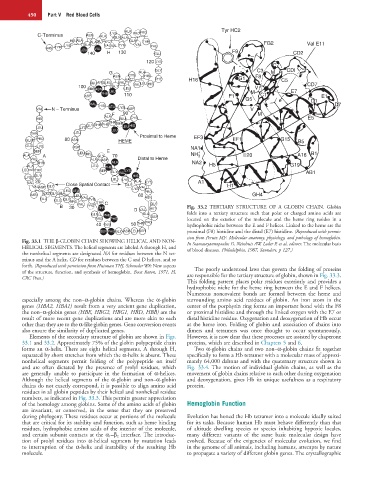
B 30 VAL PRO THR MET Fig. 33.2 TERTIARY STRUCTURE OF A GLOBIN CHAIN. Globin
D VAL
GLN PHE ALA ASP folds into a tertiary structure such that polar or charged amino acids are
ARG PHE located on the exterior of the molecule and the heme ring resides in a
40 GLU 50 THR PRO hydrophobic niche between the E and F helices. Linked to the heme are the
SER PHE GLY ASP LEU SER proximal (F8) histidine and the distal (E7) histidine. (Reproduced with permis-
sion from Perutz MF: Molecular anatomy, physiology, and pathology of hemoglobin.
Fig. 33.1 THE β-GLOBIN CHAIN SHOWING HELICAL AND NON- In Stamatoyannopoulos G, Neinhuis AW, Leder P, et al, editors: The molecular basis
HELICAL SEGMENTS. The helical segments are labeled A through H, and of blood diseases. Philadelphia, 1987, Saunders, p 127.)
the nonhelical segments are designated NA for residues between the N ter-
minus and the A helix, CD for residues between the C and D helices, and so
forth. (Reproduced with permission from Huisman THJ, Schroeder WA: New aspects The poorly understood laws that govern the folding of proteins
of the structure, function, and synthesis of hemoglobin. Boca Raton, 1971, Fl, are responsible for the tertiary structure of globin, shown in Fig. 33.3.
CRC Press.)
This folding pattern places polar residues exteriorly and provides a
hydrophobic niche for the heme ring between the E and F helices.
Numerous noncovalent bonds are formed between the heme and
especially among the non–α-globin chains. Whereas the α-globin surrounding amino acid residues of globin. An iron atom in the
genes (HBA2, HBA1) result from a very ancient gene duplication, center of the porphyrin ring forms an important bond with the F8
the non–α-globin genes (HBE, HBG2, HBG1, HBD, HBB) are the or proximal histidine and through the linked oxygen with the E7 or
result of more recent gene duplications and are more akin to each distal histidine residue. Oxygenation and deoxygenation of Hb occur
other than they are to the α-like globin genes. Gene conversion events at the heme iron. Folding of globin and association of chains into
also ensure the similarity of duplicated genes. dimers and tetramers was once thought to occur spontaneously.
Elements of the secondary structure of globin are shown in Figs. However, it is now clear that these processes are assisted by chaperone
33.1 and 33.2. Approximately 75% of the globin polypeptide chain proteins, which are described in Chapters 5 and 6.
forms an α-helix. There are eight helical segments, A through H, Two α-globin chains and two non–α-globin chains fit together
separated by short stretches from which the α-helix is absent. These specifically to form a Hb tetramer with a molecular mass of approxi-
nonhelical segments permit folding of the polypeptide on itself mately 64,000 daltons and with the quaternary structure shown in
and are often dictated by the presence of prolyl residues, which Fig. 33.4. The motion of individual globin chains, as well as the
are generally unable to participate in the formation of α-helices. movement of globin chains relative to each other during oxygenation
Although the helical segments of the α-globin and non–α-globin and deoxygenation, gives Hb its unique usefulness as a respiratory
chains do not exactly correspond, it is possible to align amino acid protein.
residues in all globin peptides by their helical and nonhelical residue
numbers, as indicated in Fig. 33.3. This permits greater appreciation
of the homology among globins. Some of the amino acids of globin Hemoglobin Function
are invariant, or conserved, in the sense that they are preserved
during phylogeny. These residues occur at portions of the molecule Evolution has honed the Hb tetramer into a molecule ideally suited
that are critical for its stability and function, such as heme binding for its tasks. Because human Hb must behave differently than that
residues, hydrophobic amino acids of the interior of the molecule, of altitude dwelling species or species inhabiting hypoxic locales,
and certain subunit contacts at the α 1–β 2 interface. The introduc- many different variants of the same basic molecular design have
tion of prolyl residues into α-helical segments by mutation leads evolved. Because of the exigencies of molecular evolution, we find
to interruption of the α-helix and instability of the resulting Hb in the genome of all animals, including humans, attempts by nature
molecule. to propagate a variety of different globin genes. The crystallographic