Page 1192 - Williams Hematology ( PDFDrive )
P. 1192
1166 Part IX: Lymphocytes and Plasma Cells Chapter 75: Functions of B Lymphocytes and Plasma Cells in Immunoglobulin Production 1167
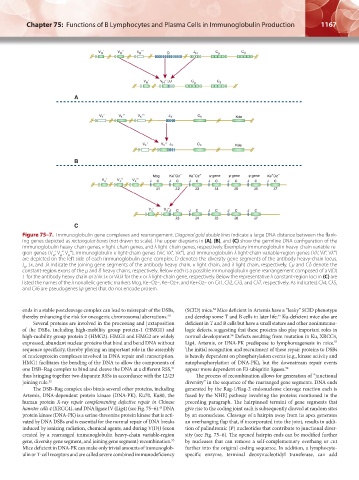
Figure 75–7. Immunoglobulin gene complexes and rearrangement. Diagonal gold double lines indicate a large DNA distance between the flank-
ing genes depicted as rectangular boxes (not drawn to scale). The upper diagrams in (A), (B), and (C) show the germline DNA configuration of the
immunoglobulin heavy-chain genes, κ light-chain genes, and λ light-chain genes, respectively. Exemplary immunoglobulin heavy-chain variable-re-
gion genes (V ‘, V ‘‘, V ‘‘‘), immunoglobulin κ light-chain genes (Vκ‘, Vκ‘‘, Vκ‘‘‘), and immunoglobulin λ light-chain variable-region genes (Vλ‘, Vλ‘‘, Vλ‘‘‘)
H
H
H
are depicted on the left side of each immunoglobulin gene complex. D denotes the diversity gene segments of the antibody heavy-chain locus.
J , Jκ, and Jλ indicate the joining gene segments of the antibody heavy chain, κ light chain, and λ light chain, respectively. Cμ and Cδ denote the
H
constant-region exons of the μ and δ heavy chains, respectively. Below each is a possible immunoglobulin gene rearrangement composed of a V(D)
J for the antibody heavy chain or a Vκ Jκ or VλJλ for the κ or λ light-chain gene, respectively. Below the representative λ constant-region loci in (C) are
listed the names of the λ nonallelic genetic markers Mcg, Ke–Oz–, Ke–Oz+, and Ke+Oz– on Cλ1, Cλ2, Cλ3, and Cλ7, respectively. As indicated, Cλ4, Cλ5,
and Cλ6 are pseudogenes (ψ gene) that do not encode protein.
54
ends in a stable postcleavage complex can lead to misrepair of the DSBs, (SCID) mice. Mice deficient in Artemis have a “leaky” SCID phenotype
55
thereby enhancing the risk for oncogenic chromosomal aberrations. 50 and develop some T and B cells in later life. Ku-deficient mice also are
Several proteins are involved in the processing and juxtaposition deficient in T and B cells but have a small stature and other nonimmuno-
of the DSBs, including high-mobility group protein-1 (HMG1) and logic defects, suggesting that these proteins also play important roles in
56
high-mobility group protein 2 (HMG2). HMG1 and HMG2 are widely normal development. Defects resulting from mutation in Ku, XRCC4,
expressed, abundant nuclear proteins that bind and bend DNA without Lig4, Artemis, or DNA-PK predispose to lymphomagenesis in mice.
57
sequence specificity, thereby playing an important role in the assembly The initial recognition and recruitment of these repair proteins to DSBs
of nucleoprotein complexes involved in DNA repair and transcription. is heavily dependent on phosphorylation events (e.g., kinase activity and
HMG1 facilitates the bending of the DNA to allow the components of autophosphorylation of DNA-PK), but the downstream repair events
one DSB–Rag complex to bind and cleave the DNA at a different RSS, appear more dependent on E3-ubiquitin ligases. 58
51
thus bringing together two disparate RSSs in accordance with the 12/23 The process of recombination allows for generation of “junctional
joining rule. 52 diversity” in the sequence of the rearranged gene segments. DNA ends
The DSB–Rag complex also binds several other proteins, including generated by the Rag-1/Rag-2 endonuclease cleavage reaction each is
Artemis, DNA-dependent protein kinase (DNA-PK), Ku70, Ku80, the fused by the NHEJ pathway involving the proteins mentioned in the
human protein X-ray repair complementing defective repair in Chinese preceding paragraph. The hairpinned termini of gene segments that
43
hamster cells 4 (XRCC4), and DNA ligase IV (Lig4) (see Fig. 75–6). DNA give rise to the coding joint each is subsequently cleaved at random sites
protein kinase (DNA-PK) is a serine-threonine protein kinase that is acti- by an exonuclease. Cleavage of a hairpin away from its apex generates
vated by DNA DSBs and is essential for the normal repair of DNA breaks an overhanging flap that, if incorporated into the joint, results in addi-
induced by ionizing radiation, chemical agents, and during V(D)J (exon tion of palindromic (P) nucleotides that contribute to junctional diver-
created by a rearranged immunoglobulin heavy-chain variable-region sity (see Fig. 75–6). The opened hairpin ends can be modified further
gene, diversity gene segment, and joining gene segment) recombination. by nucleases that can remove a self-complementary overhang or cut
53
Mice deficient in DNA-PK can make only trivial amounts of immunoglob- further into the original coding sequence. In addition, a lymphocyte-
ulin or T-cell receptors and are called severe combined immunodeficiency specific enzyme, terminal deoxynucleotidyl transferase, can add
Kaushansky_chapter 75_p1159-1174.indd 1167 9/21/15 12:11 PM