Page 1191 - Williams Hematology ( PDFDrive )
P. 1191
1166 Part IX: Lymphocytes and Plasma Cells Chapter 75: Functions of B Lymphocytes and Plasma Cells in Immunoglobulin Production 1167
thus deleting the κ light-chain constant-region exon. Many of the
38
proximal IGKV genes in the so-named p region are in the opposite ori-
entation of the IGKJ segments, requiring that the V exons in this region
undergo inversion during immunoglobulin gene rearrangement. A
650-bp sequence corresponding to DNase I hypersensitive sites HS1–2
within the IGKV-IGKJ intervening region binds a CCCTC-binding fac-
tor, which directs locus contraction and long-range IGKV gene usage;
its deletion results in a sevenfold increase in proximal IGKV gene usage
along with approximately 50 percent reduction in overall locus con-
39
traction. Subsequent to κ light-chain gene rearrangement, one of the
41 functional Vλ exons can rearrange with any one of the four func-
tional JλCλ exons to generate a gene that can encode a λ light chain
(Fig. 75–7C).
Somatic V-region gene recombination involves introduction of
double-strand DNA breaks at RSS, juxtaposition of the broken ends,
and then religation through a process called nonhomologous DNA
end-joining (NHEJ). The first cleavage step requires a specialized het-
erodimeric endonuclease complex comprised of Rag-1 and Rag-2 (see
Fig. 75–6). Rag-1 and Rag-2 are encoded by adjacent genes located on
the short arm of chromosome 11 (11p13–p12). Mice with either RAG
gene knocked out cannot undergo immunoglobulin or T-cell receptor
gene rearrangements and consequently fail to produce mature B or
T lymphocytes. Mutations that impair, but do not completely abolish,
40
the function of Rag-1 or Rag-2 in humans result in a form of combined
immune deficiency termed Omenn syndrome. 41
The process of somatic DNA recombination is initiated when the
Rag (recombination activating gene) endonuclease introduces DNA
double-strand breaks (DSBs) at the border of two recombining gene
42
segments and their flanking RSSs. DNA cleavage by Rag leads to four
broken DNA ends that are repaired and joined through a process called
NHEJ to form coding and signal joints. 43,44 Occasionally these DSBs can
be repaired aberrantly, leading to the formation of chromosomal lesions
such as translocations, deletions, or inversions, 45,46 often found in B-cell
neoplasms. If the breakpoints of these chromosomal lesions lie near
potential oncogenes or tumor-suppressor genes, they can lead to cellu-
lar transformation and lymphoid tumors. The mechanism of DNA rear-
rangement is similar for the heavy- and light-chain loci. However, only
one joining event is needed to generate a light-chain gene, whereas two
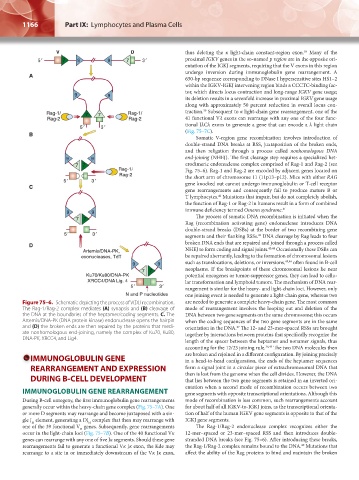
Figure 75–6. Schematic depicting the process of V(D)J recombination. are needed to generate a complete heavy-chain gene. The most common
The Rag-1/Rag-2 complex mediates (A) synapsis and (B) cleavage of mode of rearrangement involves the looping out and deletion of the
the DNA at the boundaries of the heptamer/coding segments. C. The DNA between two gene segments on the same chromosome; this occurs
Artemis/DNA-PK (DNA protein kinase) endonuclease opens the hairpin when the coding sequences of the two gene segments are in the same
and (D) the broken ends are then repaired by the proteins that medi- orientation in the DNA. The 12- and 23-mer-spaced RSSs are brought
47
ate nonhomologous end-joining, namely the complex of Ku70, Ku80, together by interactions between proteins that specifically recognize the
DNA-PK, XRCC4, and Lig4.
length of the spacer between the heptamer and nonamer signals, thus
accounting for the 12/23 joining rule. 36,48 The two DNA molecules then
IMMUNOGLOBULIN GENE are broken and rejoined in a different configuration. By joining precisely
in a head-to-head configuration, the ends of the heptamer sequences
REARRANGEMENT AND EXPRESSION form a signal joint in a circular piece of extrachromosomal DNA that
DURING B-CELL DEVELOPMENT then is lost from the genome when the cell divides. However, the DNA
that lies between the two gene segments is retained in an inverted ori-
entation when a second mode of recombination occurs between two
IMMUNOGLOBULIN GENE REARRANGEMENT gene segments with opposite transcriptional orientations. Although this
During B-cell ontogeny, the first immunoglobulin gene rearrangements mode of recombination is less common, such rearrangements account
generally occur within the heavy-chain gene complex (Fig. 75–7A). One for about half of all IGKV-to-IGKJ joins, as the transcriptional orienta-
or more D segments may rearrange and become juxtaposed with a sin- tion of half of the human IGKV gene segments is opposite to that of the
gle J element, generating a DJ complex that then may rearrange with IGKJ gene segments.
H
H
one of the 39 functional V genes. Subsequently, gene rearrangements The Rag-1/Rag-2 endonuclease complex recognizes either the
H
occur in the light-chain loci (Fig. 75–7B). One of the 40 functional Vκ 12-mer–spaced or 23-mer–spaced RSS and then introduces double-
genes can rearrange with any one of five Jκ segments. Should these gene stranded DNA breaks (see Fig. 75–6). After introducing these breaks,
49
rearrangements fail to generate a functional Vκ Jκ exon, the Kde may the Rag-1/Rag-2 complex remains bound to the DNA. Mutations that
rearrange to a site in or immediately downstream of the Vκ Jκ exon, affect the ability of the Rag proteins to bind and maintain the broken
Kaushansky_chapter 75_p1159-1174.indd 1166 9/21/15 12:11 PM