Page 181 - Williams Hematology ( PDFDrive )
P. 181
156 Part IV: Molecular and Cellular Hematology Chapter 11: Genomics 157
O
Genomic DNA
HN Cleavage
site Fluor
3’ 5’ O N
Fragment (200–500 bp)
PPP O
3
Block
Ligate adapters
A1 SP1 A Emission
T
G Incorporate
SP2 A2 C G C T Detect
Generate clusters T A Deblock
C
SP2 A2 C G A Cleave fluor
A T T Excitation
A
Flowcell G C
SP1 A1 C
A C O
G X
T A HN
T 5
SP1 Sequence first end G C DNA O N
G
A2 A
C O O
5’
3
Regerate clusters and
SP2 sequence paired end OH free 3 end
A1
A B
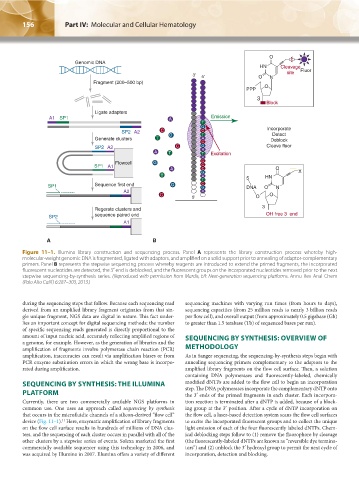
Figure 11–1. Illumina library construction and sequencing process. Panel A represents the library construction process whereby high-
molecular-weight genomic DNA is fragmented, ligated with adaptors, and amplified on a solid support prior to annealing of adaptor-complementary
primers. Panel B represents the stepwise sequencing process whereby reagents are introduced to extend the primed fragments, the incorporated
fluorescent nucleotides are detected, the 3′ end is deblocked, and the fluorescent groups on the incorporated nucleotides removed prior to the next
stepwise sequencing-by-synthesis series. (Reproduced with permission from Mardis, ER: Next-generation sequencing platforms. Annu Rev Anal Chem
(Palo Alto Calif) 6:287–303, 2013.)
during the sequencing steps that follow. Because each sequencing read sequencing machines with varying run times (from hours to days),
derived from an amplified library fragment originates from that sin- sequencing capacities (from 25 million reads to nearly 3 billion reads
gle unique fragment, NGS data are digital in nature. This fact under- per flow cell), and overall output (from approximately 0.5 gigabase (Gb)
lies an important concept for digital sequencing methods: the number to greater than 1.5 terabase (Tb) of sequenced bases per run).
of specific sequencing reads generated is directly proportional to the
amount of input nucleic acid, accurately reflecting amplified regions of SEQUENCING BY SYNTHESIS: OVERVIEW OF
a genome, for example. However, as the generation of libraries and the
amplification of fragments involve polymerase chain reaction (PCR) METHODOLOGY
amplification, inaccuracies can result via amplification biases or from As in Sanger sequencing, the sequencing-by-synthesis steps begin with
PCR enzyme substitution errors in which the wrong base is incorpo- annealing sequencing primers complementary to the adaptors to the
rated during amplification. amplified library fragments on the flow cell surface. Then, a solution
containing DNA polymerases and fluorescently-labeled, chemically
SEQUENCING BY SYNTHESIS: THE ILLUMINA modified dNTPs are added to the flow cell to begin an incorporation
step. The DNA polymerases incorporate the complementary dNTP onto
PLATFORM the 3′ ends of the primed fragments in each cluster. Each incorpora-
Currently, there are two commercially available NGS platforms in tion reaction is terminated after a dNTP is added, because of a block-
common use. One uses an approach called sequencing by synthesis ing group at the 3′ position. After a cycle of dNTP incorporation on
that occurs in the microfluidic channels of a silicon-derived “flow cell” the flow cell, a laser-based detection system scans the flow cell surfaces
device (Fig. 11–1). Here, enzymatic amplification of library fragments to excite the incorporated fluorescent groups and to collect the unique
11
on the flow cell surface results in hundreds of millions of DNA clus- light emission of each of the four fluorescently labeled dNTPs. Chem-
ters, and the sequencing of each cluster occurs in parallel with all of the ical deblocking steps follow to (1) remove the fluorophore by cleavage
other clusters by a stepwise series of events. Solexa marketed the first (the fluorescently-labeled dNTPs are known as “reversible dye termina-
commercially available sequencer using this technology in 2006, and tors”) and (2) unblock the 3′ hydroxyl group to permit the next cycle of
was acquired by Illumina in 2007. Illumina offers a variety of different incorporation, detection and blocking.
Kaushansky_chapter 11_p0155-0164.indd 156 9/18/15 11:48 PM