Page 192 - Williams Hematology ( PDFDrive )
P. 192
166 Part IV: Molecular and Cellular Hematology Chapter 12: Epigenetics 167
B lymphocytes. Furthermore, the recruitment of the ISWI-family com- being acetylation, methylation, ubiquitylation, and phosphorylation.
10
plex nucleosome remodeling factor (NURF) to the early growth response An inventory and functional analysis of all of these modifications, the
protein 1 (EGR1) locus (important for thymocyte maturation) involves enzymes that place and remove these modifications, is beyond the scope
interaction with the transcription factor serum response factor (SRF) of this chapter; however, more important are the concepts, which can
11
by the NURF subunit BPTF, enabling its stable binding to promoters. then be applied widely to various contexts.
Notably, Ikaros (which drives lymphoid differentiation) acts to inhibit First, the vast majority of histone modifications occur either on
both the ATP-dependent remodeling and HDAC activities of NuRD at the extended aminoterminal “tails” of histones, whereas a minority also
13
target genes to enable activation rather than silencing. Taken together, occur on the histone octamer “core.” The core of the histone octamer
12
these and other examples illustrate the use of remodeler function and wraps the DNA, whereas histone tails serve as platforms for the regu-
recruitment to activate or repress key genes in blood differentiation. lated binding of proteins, and covalent modifications can either enhance
or deter binding of chromatin remodelers, chromatin modifiers, and
transcription factors, and help to orchestrate protein associations dur-
PRINCIPLES OF HISTONE ing transcription (Fig. 12–2). For example, methylation on histone H3
MODIFICATION (H3) H3K4me can deter interaction with DNMTs and therefore cause
passive DNA demethylation; in contrast, H3K4me3 can facilitate inter-
14
HISTONE MODIFICATION CONCEPTS: WRITE, action with RNA polymerase II (RNAP II) machinery. Second, histone
READ, ERASE modifiers are typically targeted by site-specific DNA binding proteins
(see Fig. 12–2), which are themselves responsive to developmental and
The process of transcriptional regulation is accompanied by the environmental/metabolic signaling. Third, some histone modifiers are
ordered placement of particular histone modifications at enhancers, targeted or regulated by other histone modifications, which underlies
promoters, and coding regions. There are dozens of different modifi- (in part) why certain sets of histone modifications are coincident in
cations that occur on histones, with the most common modifications regions. 13
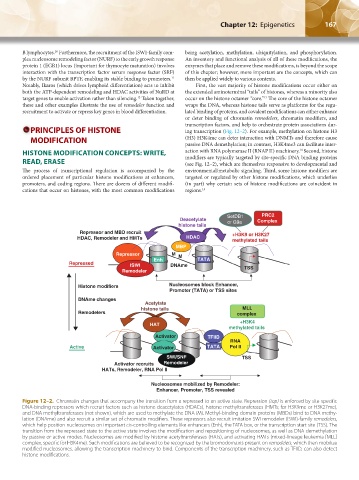
SetDB1 PRC2
Deacetylate or G9a Complex
histone tails
Repressor and MBD recruit +H3K9 or H3K27
HDAC, Remodeler and HMTs HDAC methylated tails
MBP
Repressor M M
Enh
Repressed ISWI Enh DNAme TATA
Remodeler TSS
Histone modifiers Nucleosomes block Enhancer,
Promoter (TATA) or TSS sites
DNAme changes
Acetylate
histone tails MLL
Remodelers complex
+H3K4
HAT
methylated tails
Activator TFIID
RNA
Active Activator TATA Pol II
SWI/SNF TSS
Activator recruits Remodeler
HATs, Remodeler, RNA Pol II
Nucleosomes mobilized by Remodeler:
Enhancer, Promoter, TSS revealed
Figure 12–2. Chromatin changes that accompany the transition from a repressed to an active state. Repression (top) is enforced by site-specific
DNA-binding repressors which recruit factors such as histone deacetylates (HDACs), histone methyltransferases (HMTs; for H3K9me or H3K27me),
and DNA methyltransferases (not shown), which are used to methylate the DNA (M). Methyl-binding domain proteins (MBDs) bind to DNA methy-
lation (DNAme) and also recruit a similar set of chromatin modifiers. These repressors also recruit imitation SWI remodeler (ISWI)-family remodelers,
which help position nucleosomes on important cis-controlling elements like enhancers (Enh), the TATA box, or the transcription start site (TSS). The
transition from the repressed state to the active state involves the modification and repositioning of nucleosomes, as well as DNA demethylation
by passive or active modes. Nucleosomes are modified by histone acetyltransferases (HATs), and activating HMTs (mixed-lineage leukemia [MLL]
complex, specific for H3K4me). Such modifications are believed to be recognized by the bromodomains present on remodelers, which then mobilize
modified nucleosomes, allowing the transcription machinery to bind. Components of the transcription machinery, such as TFIID, can also detect
histone modifications.
Kaushansky_chapter 12_p0165-0172.indd 167 17/09/15 6:28 pm