Page 1943 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 1943
Chapter 113 Human Leukocyte Antigen and Human Neutrophil Antigen Systems 1723
STRUCTURE OF THE HUMAN LEUKOCYTE
ANTIGEN CLASS I AND II
α 2 The structure of HLA molecules and their relationship with their
α natural ligand, the TCR, has been well characterized by
1
crystallography. 9–11 HLA molecules are heterodimer glycoproteins
belonging to the immunoglobulin superfamily with common features
(Fig. 113.4). This includes two α-helical domains protruding toward
the extracellular milieu. Between them lies a flat surface formed by
β-sheet structures that contributes to the formation of a groove
accommodating peptides generated from intracellular (HLA class I)
or extracellular proteins (HLA class II) (Fig. 113.5). The helices/
N
peptide complex is exposed for TCR recognition. Because HLA
polymorphism is clustered within the α-helixes and β-sheets domains,
N peptides display variable affinity for distinct HLA alleles. 12,13 It has
been proposed that a given peptide can bind to closely related alleles,
and HLA superfamilies with similar binding characteristics have been
described. 14–16 However, peptide binding to related but distinct HLA
alleles is associated with conformational dissimilarity caused by dif-
17
ferential interaction with variant residues in the binding groove.
The TCR interaction with HLA required for productive engagement
spans a surface including the peptide and portions of the α- and β-
helix. 11,18–20 This double requirement of interaction between TCR
and HLA/peptide complex represents the structural basis for HLA
restriction. Degenerate and promiscuous TCR recognition of peptides
C presented by distinct HLA alleles within the same superfamily has
21
C also been described. Although this concept holds in general, several
β m exceptions can be expected because single amino acid variants in the
2
HLA molecule may disallow binding of a peptide or may not be
α 3 permissive to TCR engagement. 22–24
The binding affinity of a given peptide for an HLA allele can
be predicted through algorithms that compile available informa-
tion to identify amino acid residues that fit distinct pockets of
the HLA groove. 13,25 Several algorithms implement this informa-
tion with experimental testing based on the refolding capability
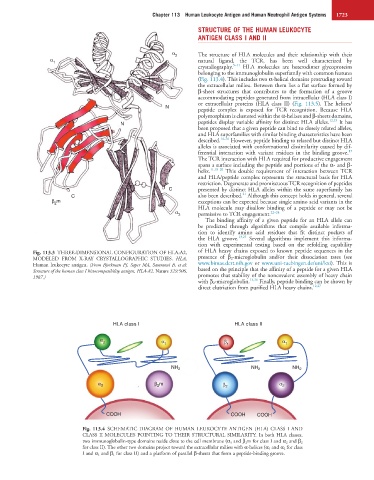
Fig. 113.3 THREE-DIMENSIONAL CONFIGURATION OF HLA-A2, of HLA heavy chains exposed to known peptide sequences in the
MODELED FROM X-RAY CRYSTALLOGRAPHIC STUDIES. HLA, presence of β 2 -microglobulin and/or their dissociation rates (see
Human leukocyte antigen. (From Bjorkman PJ, Saper MA, Samraoui B, et al: www.bimas.dcrt.nih.gov or www.uni-tuebingen.de/uni/kxi). This is
Structure of the human class I histocompatibility antigen, HLA-A2. Nature 329:506, based on the principle that the affinity of a peptide for a given HLA
1987.) promotes that stability of the noncovalent assembly of heavy chain
13,26
with β 2 -microglobulin. Finally, peptide binding can be shown by
direct elutriation from purified HLA heavy chains. 12,27
HLA class I HLA class II
α α
2 α 1 β 1 1
NH 2 NH 2 NH 2
α 3 β m β 2 α 2
2
COOH COOH COOH
Fig. 113.4 SCHEMATIC DIAGRAM OF HUMAN LEUKOCYTE ANTIGEN (HLA) CLASS I AND
CLASS II MOLECULES POINTING TO THEIR STRUCTURAL SIMILARITY. In both HLA classes,
two immunoglobulin-type domains reside close to the cell membrane (α 3 and β 2m for class I and α 2 and β 2
for class II). The other two domains project toward the extracellular milieu with α-helices (α 1 and α 2 for class
I and α 1 and β 1 for class II) and a platform of parallel β-sheets that form a peptide-binding groove.