Page 40 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 40
12 Part I Molecular and Cellular Basis of Hematology
mutations confers selective advantage in the face of unusual environ- A
mental conditions, such as malaria epidemics. These “adaptive” Hpa I β S Hpa I Hpa I Southern blot
mutations drive the dynamic change in the genome with time
(evolution). bA bS
Most of the mutations that accumulate in the DNA of Homo
sapiens occur in either intergenic DNA or the “silent” bases of DNA, 13.0 kb
such as the degenerate third bases of codons. They do not pathologi- Hpa I β A Hpa I Hpa I
cally alter the function of the gene or its products. These clinically
harmless mutations are called DNA polymorphisms. DNA polymor-
phisms can be regarded in exactly the same way as other types of 7.6 kb 6.4 kb
polymorphisms that have been widely recognized for years (e.g., eye
and hair color, blood groups). They are variations in the population
that occur without apparent clinical impact. Each of us differs from B
other humans in the precise number and type of DNA polymor- Hpa I a Hpa I Southern blots
phisms that we possess. Most polymorphisms represent single- 2 α 1 VNTR
nucleotide changes and are called single-nucleotide polymorphisms Pt#1 Patients
(SNPs). Hpa I Hpa I 1 2 3
Similar to other types of polymorphisms, DNA polymorphisms a 2 α 1 VNTR
breed true. In other words, if an individual’s DNA contains a G 1200 Pt#2
bases upstream from the α-globin gene, instead of the C most com- Hpa I a α Hpa I
monly found in the population, that G will be transmitted to that 2 1 VNTR
individual’s offspring. Note that if one had a means for distinguishing Pt#3
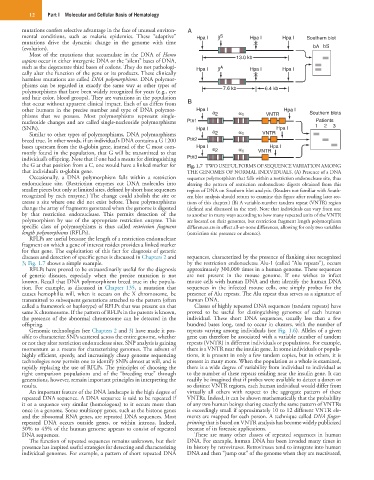
the G at that position from a C, one would have a linked marker for Fig. 1.7 TWO USEFUL FORMS OF SEQUENCE VARIATION AMONG
that individual’s α-globin gene. THE GENOMES OF NORMAL INDIVIDUALS. (A) Presence of a DNA
Occasionally, a DNA polymorphism falls within a restriction sequence polymorphism that falls within a restriction endonuclease site, thus
endonuclease site. (Restriction enzymes cut DNA molecules into altering the pattern of restriction endonuclease digests obtained from this
smaller pieces but only at limited sites, defined by short base sequences region of DNA on Southern blot analysis. (Readers not familiar with South-
recognized by each enzyme.) The change could abolish the site or ern blot analysis should return to examine this figure after reading later sec-
create a site where one did not exist before. These polymorphisms tions of this chapter.) (B) A variable-number tandem repeat (VNTR) region
change the array of fragments generated when the genome is digested (defined and discussed in the text). Note that individuals can vary from one
by that restriction endonuclease. This permits detection of the to another in many ways according to how many repeated units of the VNTR
polymorphism by use of the appropriate restriction enzyme. This are located on their genomes, but restriction fragment length polymorphism
specific class of polymorphisms is thus called restriction fragment differences are in effect all-or-none differences, allowing for only two variables
length polymorphisms (RFLPs). (restriction site presence or absence).
RFLPs are useful because the length of a restriction endonuclease
fragment on which a gene of interest resides provides a linked marker
for that gene. The exploitation of this fact for diagnosis of genetic
diseases and detection of specific genes is discussed in Chapters 2 and sequences, characterized by the presence of flanking sites recognized
3; Fig. 1.7 shows a simple example. by the restriction endonuclease Alu-1 (called “Alu repeats”), occurs
RFLPs have proved to be extraordinarily useful for the diagnosis approximately 300,000 times in a human genome. These sequences
of genetic diseases, especially when the precise mutation is not are not present in the mouse genome. If one wishes to infect
known. Recall that DNA polymorphisms breed true in the popula- mouse cells with human DNA and then identify the human DNA
tion. For example, as discussed in Chapter 135, a mutation that sequences in the infected mouse cells, one simply probes for the
causes hemophilia will, when it occurs on the X chromosome, be presence of Alu repeats. The Alu repeat thus serves as a signature of
transmitted to subsequent generations attached to the pattern (often human DNA.
called a framework or haplotype) of RFLPs that was present on that Classes of highly repeated DNA sequences (tandem repeats) have
same X chromosome. If the pattern of RFLPs in the parents is known, proved to be useful for distinguishing genomes of each human
the presence of the abnormal chromosome can be detected in the individual. These short DNA sequences, usually less than a few
offspring. hundred bases long, tend to occur in clusters, with the number of
Genomic technologies (see Chapters 2 and 3) have made it pos- repeats varying among individuals (see Fig. 1.6). Alleles of a given
sible to characterize SNPs scattered across the entire genome, whether gene can therefore be associated with a variable number of tandem
or not they alter restriction endonuclease sites. SNP analysis is gaining repeats (VNTR) in different individuals or populations. For example,
momentum as a means for characterizing genomes. The advent of there is a VNTR near the insulin gene. In some individuals or popula-
highly efficient, speedy, and increasingly cheap genome sequencing tions, it is present in only a few tandem copies, but in others, it is
technologies now permits one to identify SNPs almost at will, and is present in many more. When the population as a whole is examined,
rapidly replacing the use of RFLPs. The principles of choosing the there is a wide degree of variability from individual to individual as
right comparison populations and of the “breeding true” through to the number of these repeats residing near the insulin gene. It can
generations, however, remain important principles in interpreting the readily be imagined that if probes were available to detect a dozen or
results. so distinct VNTR regions, each human individual would differ from
An important feature of the DNA landscape is the high degree of virtually all others with respect to the aggregate pattern of these
repeated DNA sequence. A DNA sequence is said to be repeated if VNTRs. Indeed, it can be shown mathematically that the probability
it or a sequence very similar (homologous) to it occurs more than of any two human beings sharing exactly the same pattern of VNTRs
once in a genome. Some multicopy genes, such as the histone genes is exceedingly small if approximately 10 to 12 different VNTR ele-
and the ribosomal RNA genes, are repeated DNA sequences. Most ments are mapped for each person. A technique called DNA finger-
repeated DNA occurs outside genes, or within introns. Indeed, printing that is based on VNTR analysis has become widely publicized
30% to 45% of the human genome appears to consist of repeated because of its forensic applications.
DNA sequences. There are many other classes of repeated sequences in human
The function of repeated sequences remains unknown, but their DNA. For example, human DNA has been invaded many times in
presence has inspired useful strategies for detecting and characterizing its history by retroviruses. Retroviruses tend to integrate into human
individual genomes. For example, a pattern of short repeated DNA DNA and then “jump out” of the genome when they are reactivated,