Page 583 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 583
498 Part V Red Blood Cells
ALA which is under the control of erythroid-specific promoters such as
Plumboporphyria GATA1, a globin transcription factor. Whether heme inhibits import
ALA Porphobilinogen of pre-ALAS2 into the mitochondrial matrix remains to be unequivo-
X-linked hereditary cally established. Heme may possibly also prevent the accumulation
sideroblastic anemia Acute intermittent of intracellular iron by controlling the acquisition of iron from
Glycine + transferrin (see Fig. 38.2B). In addition to transport of iron from
succinyl CoA porphyria plasma to the cytosol by the transferrin receptor, a second transport
Hydroxymethylbilane step is required for mitochondrial uptake of iron. This step is fulfilled
22
Heme by mitoferrin, a member of the solute carrier 25 family of proteins
Erythropoietic Congenital located in the inner mitochondrial membrane, which, to import iron
protoporphyria porphyria into the mitochondrion, must interact both with ferrochelatase and
Protoporphyrin IX with the adenosine triphosphate (ATP)-binding cassette transporter
23
Uroporphyrinogen III ABCB10. Levels of intracellular iron regulate the translation of
Variegate porphyria ALAS2 mRNA. Cellular iron homeostasis is maintained through a
Protoporphyrinogen III Porphyria cutanea posttranscriptional regulatory mechanism, which is mediated by iron
tarda
regulatory proteins that bind to iron-responsive elements in mRNA
Coproporphyrinogen III of target genes to either increase or decrease translation. 24,25 The RNA
binding activity of iron-responsive proteins (IRP) is regulated by
Hereditary mitochondrial iron-sulfur cluster synthesis and cytosolic iron
coproporphyria levels. 26–28 When iron is available for heme synthesis, translation of
ALAS2 is allowed to proceed as a result of decreased IRP binding to
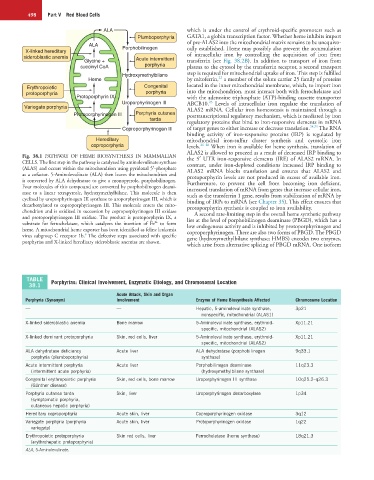
Fig. 38.1 PATHWAY OF HEME BIOSYNTHESIS IN MAMMALIAN the 5′ UTR iron-responsive elements (IRE) of ALAS2 mRNA. In
CELLS. The first step in the pathway is catalyzed by aminolevulinate synthase contrast, under iron-depleted conditions increased IRP binding to
(ALAS) and occurs within the mitochondrion using pyridoxal 5′-phosphate ALAS2 mRNA blocks translation and ensures that ALAS2 and
as a cofactor. 5-Aminolevulinate (ALA) then leaves the mitochondrion and protoporphyrin levels are not produced in excess of available iron.
is converted by ALA dehydratase to give a monopyrrole, porphobilinogen. Furthermore, to prevent the cell from becoming iron deficient,
Four molecules of this compound are converted by porphobilinogen deami- increased translation of mRNA from genes that increase cellular iron,
nase to a linear tetrapyrrole, hydroxymethylbilane. This molecule is then such as the transferrin 1 gene, results from stabilization of mRNA by
cyclized by uroporphyrinogen III synthase to uroporphyrinogen III, which is binding of IRPs to mRNA (see Chapter 35). This effect ensures that
decarboxylated to coproporphyrinogen III. This molecule enters the mito- protoporphyrin synthesis is coupled to iron availability.
chondrion and is oxidized in succession by coproporphyrinogen III oxidase A second rate-limiting step in the overall heme synthetic pathway
and protoporphyrinogen III oxidase. The product is protoporphyrin IX, a lies at the level of porphobilinogen deaminase (PBGD), which has a
2+
substrate for ferrochelatase, which catalyzes the insertion of Fe to form low endogenous activity and is inhibited by protoporphyrinogen and
heme. A mitochondrial heme exporter has been identified as feline leukemia coproporphyrinogen. There are also two forms of PBGD. The PBGD
6
virus subgroup C receptor 1b. The defective steps associated with specific gene (hydroxymethylbilane synthase; HMBS) encodes two enzymes,
porphyrias and X-linked hereditary sideroblastic anemias are shown.
which arise from alternative splicing of PBGD mRNA. One isoform
TABLE Porphyrias: Clinical Involvement, Enzymatic Etiology, and Chromosomal Location
38.1
Acute Attack, Skin and Organ
Porphyria (Synonym) Involvement Enzyme of Heme Biosynthesis Affected Chromosome Location
— — Hepatic, 5-aminolevulinate synthase, 3p21
nonspecific, mitochondrial (ALAS1)
X-linked sideroblastic anemia Bone marrow 5-Aminolevulinate synthase, erythroid- Xp11.21
specific, mitochondrial (ALAS2)
X-linked dominant protoporphyria Skin, red cells, liver 5-Aminolevulinate synthase, erythroid- Xp11.21
specific, mitochondrial (ALAS2)
ALA dehydratase deficiency Acute liver ALA dehydratase (porphobilinogen 9q33.1
porphyria (plumboporphyria) synthase)
Acute intermittent porphyria Acute liver Porphobilinogen deaminase 11q23.3
(intermittent acute porphyria) (hydroxymethylbilane synthase)
Congenital erythropoietic porphyria Skin, red cells, bone marrow Uroporphyrinogen III synthase 10q25.2–q26.3
(Günther disease)
Porphyria cutanea tarda Skin, liver Uroporphyrinogen decarboxylase 1p34
(symptomatic porphyria,
cutaneous hepatic porphyria)
Hereditary coproporphyria Acute skin, liver Coproporphyrinogen oxidase 3q12
Variegate porphyria (porphyria Acute skin, liver Protoporphyrinogen oxidase 1q22
variegata)
Erythropoietic protoporphyria Skin red cells, liver Ferrochelatase (heme synthase) 18q21.3
(erythrohepatic protoporphyria)
ALA, 5-Aminolevulinate.