Page 59 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 59
Chapter 3 Genomic Approaches to Hematology 31
1 the relative contribution of individual genetic subclones to the entire
Y
tumor, these approaches usually have limited resolution and are not
X
22
able to detect very small subclones that comprise few tumor cells.
21
2 Several novel technologies have been developed that allow DNA and
20
RNA sequencing of single tumor cells as well as of single cells from
19
the tumor microenvironment. Advances in microfluidic approaches
18
make it possible to generate RNA sequencing data from thousands
3 of cells simultaneously, and several different technological concepts
17
have emerged for highly parallel sample preparation. For example,
the entire sample processing workflow, from isolating single cells to
16
generating sequencing libraries, can be generated in multiwell plates
in order to perform multiple reactions at the same time. This can be
15 4 done with typical multiwell plates, or the entire experimental work-
flow can take place in a fully integrated microfluidic “lab on a chip.”
14 Another approach uses microdroplet technology. To this end, a single
cell is packaged into an emulsion droplet, and thousands of droplets
5 are generated. Although the single cells are segregated into individual
13 droplets, a molecular barcoding step takes place with which every
RNA molecule in each cell is labeled with a unique molecular tag.
After this step, the droplets are dissolved and RNA sequencing prepa-
12 6 ration is performed in a single tube. The barcodes can later be used
to precisely assign each RNA molecule to the correct cell, making it
11 7 possible to determine the gene expression profile of each single cell.
These technologies currently enable sequencing of thousands of
10 9 8 single cells simultaneously, but that number will undoubtedly increase
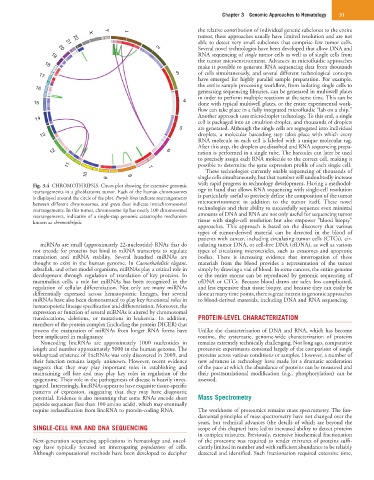
Fig. 3.4 CHROMOTHRIPSIS. Circos plot showing the extensive genomic with rapid progress in technology development. Having a methodol-
rearrangements in a glioblastoma tumor. Each of the human chromosomes ogy in hand that allows RNA sequencing with single-cell resolution
is displayed around the circle of the plot. Purple lines indicate rearrangements is particularly useful to precisely define the composition of the tumor
between different chromosomes, and green lines indicate intrachromosomal microenvironment in addition to the tumor itself. These novel
rearrangements. In this tumor, chromosome 1p has nearly 100 chromosomal technologies and their ability to successfully sequence even minimal
rearrangements, indicative of a single-step genomic catastrophe mechanism amounts of DNA and RNA are not only useful for sequencing tumor
known as chromothripsis. tissue with single-cell resolution but also empower “blood biopsy”
approaches. This approach is based on the discovery that various
types of tumor-derived material can be detected in the blood of
patients with cancer, including circulating tumor cells (CTCs), cir-
miRNAs are small (approximately 22-nucleotide) RNAs that do culating tumor DNA, or cell-free DNA (cfDNA), as well as various
not encode for proteins but bind to mRNA transcripts to regulate types of circulating microvesicles, such as exosomes and apoptotic
translation and mRNA stability. Several hundred miRNAs are bodies. There is increasing evidence that interrogation of these
thought to exist in the human genome. In Caenorhabditis elegans, materials from the blood provides a representation of the tumor
zebrafish, and other model organisms, miRNAs play a critical role in simply by drawing a vial of blood. In some cancers, the entire genome
development through regulation of translation of key proteins. In or the entire exome can be reproduced by genomic sequencing of
mammalian cells, a role for miRNAs has been recognized in the cfDNA or CTCs. Because blood draws are safer, less complicated,
regulation of cellular differentiation. Not only are many miRNAs and less expensive than tissue biopsy, and because they can easily be
differentially expressed across hematopoietic lineages, but several done at many time points, there is great interest in genomic approaches
miRNAs have also been demonstrated to play key functional roles in to blood-derived materials, including DNA and RNA sequencing.
hematopoietic lineage specification and differentiation. Moreover, the
expression or function of several miRNAs is altered by chromosomal
translocations, deletions, or mutations in leukemia. In addition, PROTEIN-LEVEL CHARACTERIZATION
members of the protein complex (including the protein DICER) that
process the maturation of miRNAs from longer RNA forms have Unlike the characterization of DNA and RNA, which has become
been implicated in malignancy. routine, the systematic, genome-wide characterization of proteins
Noncoding lincRNAs are approximately 1000 nucleotides in remains extremely technically challenging. Not long ago, comparative
length and number approximately 5000 in the human genome. The proteomic experiments consisted largely of the comparison of single
widespread existence of lincRNAs was only discovered in 2009, and proteins across various conditions or samples. However, a number of
their function remains largely unknown. However, recent evidence new advances in technology have made for a dramatic acceleration
suggests that they may play important roles in establishing and of the pace at which the abundance of proteins can be measured and
maintaining cell fate and may play key roles in regulation of the their posttranslational modification (e.g., phosphorylation) can be
epigenome. Their role in the pathogenesis of disease is heavily inves- assessed.
tigated. Interestingly, lincRNAs appear to have exquisite tissue-specific
patterns of expression, suggesting that they may have diagnostic
potential. Evidence is also mounting that some RNAs encode short Mass Spectrometry
peptide sequences (less than 100 amino acids), which may eventually
require reclassification from lincRNA to protein-coding RNA. The workhorse of proteomics remains mass spectrometry. The fun-
damental principles of mass spectrometry have not changed over the
years, but technical advances (the details of which are beyond the
SINGLE-CELL RNA AND DNA SEQUENCING scope of this chapter) have led to increased ability to detect proteins
in complex mixtures. Previously, extensive biochemical fractionation
Next-generation sequencing applications in hematology and oncol- of the proteome was required to render mixtures of proteins suffi-
ogy have typically focused on interrogating populations of cells. ciently limited in number and with sufficient abundance to be reliably
Although computational methods have been developed to decipher detected and identified. Such fractionation required extensive time,