Page 61 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 61
Chapter 3 Genomic Approaches to Hematology 33
Cell lines
Ovarian
Colon
Pancreas
Esophageal
Lung NSCLC
GBM
Lung SCLC
Melanoma
Meningioma
Breast
Gastric
Renal cell carcinoma
Others
shRNAs
Normalized
fold enrichment
3
0
–3
A
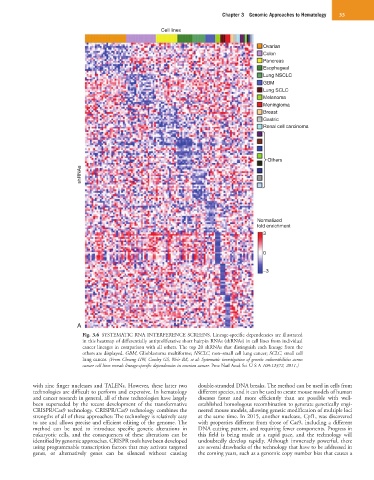
Fig. 3.6 SYSTEMATIC RNA INTERFERENCE SCREENS. Lineage-specific dependencies are illustrated
in this heatmap of differentially antiproliferative short hairpin RNAs (shRNAs) in cell lines from individual
cancer lineages in comparison with all others. The top 20 shRNAs that distinguish each lineage from the
others are displayed. GBM, Glioblastoma multiforme; NSCLC, non–small cell lung cancer; SCLC, small cell
lung cancer. (From Cheung HW, Cowley GS, Weir BA, et al: Systematic investigation of genetic vulnerabilities across
cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci U S A 108:12372, 2011.)
with zinc finger nucleases and TALENs. However, these latter two double-stranded DNA breaks. The method can be used in cells from
technologies are difficult to perform and expensive. In hematology different species, and it can be used to create mouse models of human
and cancer research in general, all of these technologies have largely diseases faster and more efficiently than are possible with well-
been superseded by the recent development of the transformative established homologous recombination to generate genetically engi-
CRISPR/Cas9 technology. CRISPR/Cas9 technology combines the neered mouse models, allowing genetic modification of multiple loci
strengths of all of these approaches: The technology is relatively easy at the same time. In 2015, another nuclease, Cpf1, was discovered
to use and allows precise and efficient editing of the genome. The with properties different from those of Cas9, including a different
method can be used to introduce specific genetic alterations in DNA cutting pattern, and requiring fewer components. Progress in
eukaryotic cells, and the consequences of these alterations can be this field is being made at a rapid pace, and the technology will
identified by genomic approaches. CRISPR tools have been developed undoubtedly develop rapidly. Although immensely powerful, there
using programmable transcription factors that may activate targeted are several drawbacks of the technology that have to be addressed in
genes, or alternatively genes can be silenced without causing the coming years, such as a genomic copy number bias that causes a