Page 608 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 608
Chapter 39 Megaloblastic Anemias 517
Via intermediates of
one-carbon metabolism
Cytoplasm
m Polyglutamation
e s
t
5-CH H PteGlu h y H PteGlu
Plasma 3 4 i n 4
membrane o t
TC II degradation
n h
Cob(I)alamin i MeCbI G
TCII E n a
TC II receptor
TC II F C,D SAH e s
Cob(III)alamin Cob(III)alamin Cob(II)alamin e
Cob(III)alamin J Reductase SAM
Methionine Homocysteine
Lysosome
Receptor-
mediated Cob(II)alamin
endocytosis
Reductase A
Mitochondria
Cob(I)alamin
Adenosyltransferase B
AdoCbl
Methylmalonyl CoA Succinyl CoA
5-CH -H PteGlu,
4
3
5-methyl-
tetrahydrofolate;
MMCoA
mutase
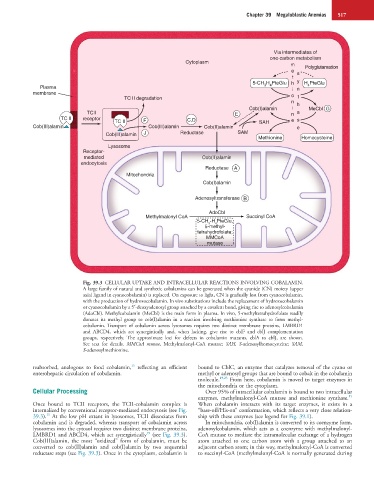
Fig. 39.3 CELLULAR UPTAKE AND INTRACELLULAR REACTIONS INVOLVING COBALAMIN.
A large family of natural and synthetic cobalamins can be generated when the cyanide (CN) moiety (upper
axial ligand in cyanocobalamin) is replaced. On exposure to light, CN is gradually lost from cyanocobalamin,
with the production of hydroxocobalamin. In vivo substitutions include the replacement of hydroxocobalamin
or cyanocobalamin by a 5′-deoxyadenosyl group attached by a covalent bond, giving rise to adenosylcobalamin
(AdoCbl). Methylcobalamin (MeCbl) is the main form in plasma. In vivo, 5-methyltetrahydrofolate readily
donates its methyl group to cob(I)alamin in a reaction involving methionine synthase to form methyl-
cobalamin. Transport of cobalamin across lysosomes requires two distinct membrane proteins, LMBRD1
and ABCD4, which act synergistically and, when lacking, give rise to cblF and cblJ complementation
groups, respectively. The approximate loci for defects in cobalamin mutants, cblA to cblJ, are shown.
See text for details. MMCoA mutase, Methylmalonyl-CoA mutase; SAH, S-adenosylhomocysteine; SAM,
S-adenosylmethionine.
15
reabsorbed, analogous to food cobalamin, reflecting an efficient bound to CblC, an enzyme that catalyzes removal of the cyano or
enterohepatic circulation of cobalamin. methyl or adenosyl groups that are bound to cobalt in the cobalamin
molecule. 19,23 From here, cobalamin is moved to target enzymes in
the mitochondria or the cytoplasm.
Cellular Processing Over 95% of intracellular cobalamin is bound to two intracellular
15
enzymes, methylmalonyl-CoA mutase and methionine synthase.
Once bound to TCII receptors, the TCII-cobalamin complex is When cobalamin interacts with its target enzymes, it exists in a
internalized by conventional receptor-mediated endocytosis (see Fig. “base-off/His-on” conformation, which reflects a very close relation-
15
39.3). At the low pH extant in lysosomes, TCII dissociates from ship with these enzymes (see legend for Fig. 39.1).
cobalamin and is degraded, whereas transport of cobalamin across In mitochondria, cob(I)alamin is converted to its coenzyme form,
lysosomes into the cytosol requires two distinct membrane proteins, adenosylcobalamin, which acts as a coenzyme with methylmalonyl-
19
LMBRD1 and ABCD4, which act synergistically (see Fig. 39.3). CoA mutase to mediate the intramolecular exchange of a hydrogen
Cob(III)alamin, the most “oxidized” form of cobalamin, must be atom attached to one carbon atom with a group attached to an
converted to cob(II)alamin and cob(I)alamin by two sequential adjacent carbon atom; in this way, methylmalonyl-CoA is converted
reductase steps (see Fig. 39.3). Once in the cytoplasm, cobalamin is to succinyl-CoA (methylmalonyl-CoA is normally generated during