Page 90 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 90
Chapter 6 Protein Architecture: Relationship of Form and Function 61
are major structural components of many cell surface and secreted The Domain Structure of Proteins
proteins, and also of many viral proteins. Protein methylation on
arginine or lysine residues is carried out by methyltransferases with In general, the minimal biologically functional unit of protein three-
S-adenosyl methionine (SAM) as the primary methyl group donor. dimensional structure is the protein domain. Domains are locally
Methylation is an important mechanism of epigenetic regulation, as compact and semi-independent units of usually contiguous polypep-
histone methylation and demethylation influence the availability of tide chain. The common size of a domain is between 100 and 200
DNA for transcription. N-acetylation, the transfer of an acetyl group amino acid residues, although much larger and smaller domains are
to the amine nitrogen at the N-terminus of the polypeptide chain, also frequently observed. Protein domains are composed of closely
occurs in a majority of eukaryotic proteins. Lysine acetylation and packed secondary structure elements—α-helices, β-sheets, or a
deacetylation is an important regulatory mechanism in a number of combination of both—and the loops that connect them. Domains
proteins. It is best characterized in histones, where histone acetyl are stabilized by hydrophobic interactions among these elements and
transferases (HATs) and histone deacetylases (HDACs) regulate gene typically have very hydrophobic central cores, with more hydrophilic
expression via modification of histone tails. Many cytoplasmic pro- amino acids extending from their surface. Alternating patterns of
teins are also acetylated, and therefore acetylation seems to play a hydrophobic residues in secondary structure elements are a reflection
2
greater role in cell biology than simply transcriptional regulation. of the role of hydrophobicity in driving protein folding and stability.
Lipidation is a modification that targets proteins to membranes in Helices are often amphipathic and pack in a folded domain such that
organelles, vesicles, and the plasma membrane. Examples of lipida- their hydrophobic face is buried in the domain interior and their
tion include myristoylation, palmitoylation, and prenylation. Each type hydrophilic face is exposed on the surface. Likewise, β-sheets often
of modification gives proteins distinct membrane affinities, although have a buried hydrophobic face and an exposed hydrophilic face. The
all types of lipidation increase the hydrophobicity of a protein and importance of the hydrophobic core to the stability of protein
thus its affinity for membranes. In N-myristoylation, the myristoyl domains is highlighted by the fact that point mutations that introduce
group (14-carbon saturated fatty acid) is transferred to an N-terminal polar or charged residues into the protein interior often cause mis-
glycine by N-myristoyltransferase. The myristoyl group does not folding and thus a loss of function. Although these general charac-
always permanently anchor the protein in the membrane; in a teristics are shared by protein domains that are found in an aqueous
number of proteins the N-terminal myristoyl group has been environment, such as that on the cytosol or on the cell surface,
observed to pack into the protein core. N-myristoylation can there- membrane-embedded proteins have very different properties reflec-
fore act as a conformational localization switch, in which protein tive of their residence in the lipid bilayer. Several common domain
conformational changes influence the availability of the handle for structures representing different categories with regard to their sec-
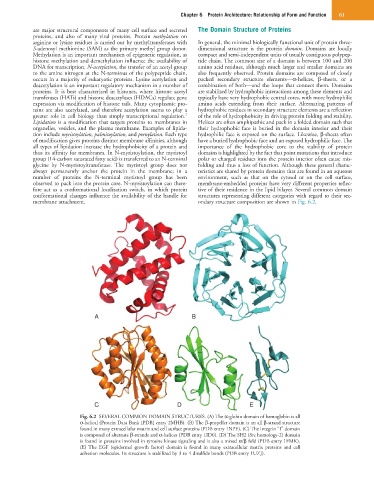
membrane attachment. ondary structure composition are shown in Fig. 6.2.
A B
C D E
Fig. 6.2 SEVERAL COMMON DOMAIN STRUCTURES. (A) The α-globin domain of hemoglobin is all
α-helical (Protein Data Bank [PDB] entry 2MHB). (B) The β-propeller domain is an all β-strand structure
found in many extracellular matrix and cell surface proteins (PDB entry 1NPE). (C) The integrin “I” domain
is composed of alternate β-strands and α-helices (PDB entry 1ID0). (D) The SH2 (Src homology-2) domain
is found in proteins involved in tyrosine kinase signaling and is also a mixed α/β fold (PDB entry 1FMK).
(E) The EGF (epidermal growth factor) domain is found in many extracellular matrix proteins and cell
adhesion molecules. Its structure is stabilized by 3 to 4 disulfide bonds (PDB entry 1UZJ).