Page 91 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 91
62 Part I Molecular and Cellular Basis of Hematology
12
Deciphering this basic protein building block is key for under- transduction. Unstructured segments are well suited for protein
standing the structure and evolution of proteins. Kinetically, the interactions controlled by posttranslational modifications. For example,
domain structure of a protein may simplify the folding process into sites of tyrosine phosphorylation are typically unstructured and
3
a step-wise course. Thus a long amino acid sequence may fold into therefore accessible for modification, but after phosphorylation they
multiple domains rapidly and correctly. For many proteins, individ- become ordered upon phosphorylation-dependent binding to a
ual domains fold in a cotranslational manner; from the N-terminal partner protein.
region, a growing nascent polypeptide chain immediately begins to Most proteins are composed of multiple domains, which may
fold domain-by-domain during translation from the ribosome in a confer multiple functions, couple a targeting function to a catalytic
4
very efficient manner. Genetically, it was long suspected that the function, or provide for allosteric regulation. In the following sections
exon structure of genes was correlated with the domain structure of we highlight the structure of a few proteins and domains and that
5
proteins. Subsequent multigenome analysis did find a strong corre- are of central and recurring importance in hematology in order to
lation between domain organization and exon–intron arrangement illustrate the relationship between domain architecture and function.
in genomic DNA. The exon–domain correlation facilitates extensive We discuss representative examples from the extracellular space (the
6
exon shuffling events during evolution, although it is not necessar- immunoglobulin domain), from intracellular signaling (protein
ily always one-exon/one-domain. This mechanism ensures that a kinase domain), and from the cell membrane (G protein–coupled
stable and functionally efficient domain can be repeatedly used as a receptors and the vitamin K receptor).
module assembled into many proteins with shared functions. A
well-known early example is the nucleotide-binding domain identi-
fied in various dehydrogenases; its robust alternate β-strand–α- The Immunoglobulin Domain and Variations
helix–β-strand fold provides a common structural unit for these
enzymes. 7 As implied by its name, the immunoglobulin (Ig) domain was first
13
Recent computational approaches demonstrate that almost all the recognized in antibodies. A detailed discussion on antibody biology
growing number of known sequences come from new combinations can be found in Chapter 24. The human genome project has identi-
of various domains, and more than 70% of all sequences can be fied the Ig superfamily (IgSF) as the largest superfamily in human
8
partially modeled from known structures with homologous domains. genome, due to its extensive usage in a more recently developed
9
9
This has been reflected in the human genome sequence. Impressive immune system in vertebrates. In fact, the Ig domain is an evolu-
progress has already been made in computational protein prediction tionarily ancient structural unit that can be found in Caenorhabditis
14
and design, principally based on the known structural elements. 10 elegans. Although Ig-like domains also exist in a few intracellular
Importantly, not all protein sequences fold into a compact domain. proteins, they are found predominantly in the extracellular space and
Depending on computational methods used, 35% to 50% of the are the most abundant structural unit found in cell surface receptors,
human proteome is estimated to lack a folded three-dimensional serving key recognition functions in both the immune and nervous
structure. Nevertheless, these intrinsically disordered proteins (or more systems. Along with a handful of other modular domains such as
often, intrinsically disordered regions within proteins) can perform fibronectin type III domains and epidermal growth factor (EGF)
critically important biologic functions that complement those of domains, they form modular structures of most receptor molecules
11
structured proteins. Intrinsically disordered protein segments typi- on the cell surface. 15
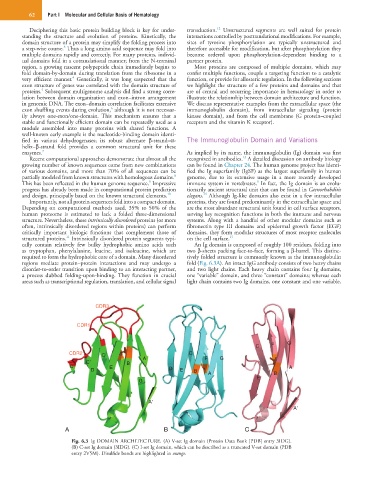
cally contain relatively few bulky hydrophobic amino acids such An Ig domain is composed of roughly 100 residues, folding into
as tryptophan, phenylalanine, leucine, and isoleucine, which are two β-sheets packing face-to-face, forming a β-barrel. This distinc-
required to form the hydrophobic core of a domain. Many disordered tively folded structure is commonly known as the immunoglobulin
regions mediate protein–protein interactions and may undergo a fold (Fig. 6.3A). An intact IgG antibody consists of two heavy chains
disorder-to-order transition upon binding to an interacting partner, and two light chains. Each heavy chain contains four Ig domains,
a process dubbed folding-upon-binding. They function in crucial one “variable” domain, and three “constant” domains; whereas each
areas such as transcriptional regulation, translation, and cellular signal light chain contains two Ig domains, one constant and one variable.
CDR3
CDR1
C F G
CDR2 C F C
C’ B A F G
C” E D A
D E B A B
G E
A’
A’
A B C
Fig. 6.3 Ig DOMAIN ARCHITECTURE. (A) V-set Ig domain (Protein Data Bank [PDB] entry 3IDG).
(B) C-set Ig domain (3IDG). (C) I-set Ig domain, which can be described as a truncated V-set domain (PDB
entry 2V5M). Disulfide bonds are highlighted in orange.