Page 1537 - Williams Hematology ( PDFDrive )
P. 1537
1512 Part XI: Malignant Lymphoid Diseases Chapter 91: Acute Lymphoblastic Leukemia 1513
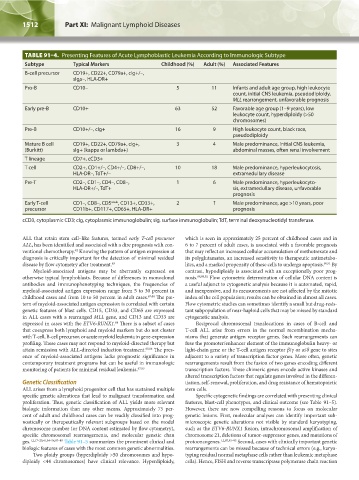
TABLE 91–4. Presenting Features of Acute Lymphoblastic Leukemia According to Immunologic Subtype
Subtype Typical Markers Childhood (%) Adult (%) Associated Features
B-cell precursor CD19+, CD22+, CD79a+, cIg+/–,
sIgμ–, HLA-DR+
Pro-B CD10– 5 11 Infants and adult age group, high leukocyte
count, initial CNS leukemia, pseudodiploidy,
MLL rearrangement, unfavorable prognosis
Early pre-B CD10+ 63 52 Favorable age group (1–9 years), low
leukocyte count, hyperdiploidy (>50
chromosomes)
Pre-B CD10+/–, cIg+ 16 9 High leukocyte count, black race,
pseudodiploidy
Mature B cell CD19+, CD22+, CD79a+, cIg+, 3 4 Male predominance, initial CNS leukemia,
(Burkitt) sIg+ (kappa or lambda+) abdominal masses, often renal involvement
T lineage CD7+, cCD3+
T cell CD2+, CD1+/–, CD4+/–, CD8+/–, 10 18 Male predominance, hyperleukocytosis,
HLA-DR–, TdT+/– extramedullary disease
Pre-T CD2–, CD1–, CD4–, CD8–, 1 6 Male predominance, hyperleukocyto-
HLA-DR+/–, TdT+ sis, extramedullary disease, unfavorable
prognosis
Early T-cell CD1–, CD8–, CD5 weak , CD13+, CD33+, 2 ? Male predominance, age >10 years, poor
precursor CD11b+, CD117+, CD65+, HLA-DR+ prognosis
cCD3, cytoplasmic CD3; cIg, cytoplasmic immunoglobulin; sIg, surface immunoglobulin; TdT, terminal deoxynucleotidyl transferase.
ALL that retain stem cell–like features, termed early T-cell precursor which is seen in approximately 25 percent of childhood cases and in
ALL, has been identified and associated with a dire prognosis with con- 6 to 7 percent of adult cases, is associated with a favorable prognosis
ventional chemotherapy. Knowing the pattern of antigen expression at that may reflect an increased cellular accumulation of methotrexate and
82
diagnosis is critically important for the detection of minimal residual its polyglutamates, an increased sensitivity to therapeutic antimetabo-
disease by flow cytometry after treatment. 83 lites, and a marked propensity of these cells to undergo apoptosis. 90,91 By
Myeloid-associated antigens may be aberrantly expressed on contrast, hypodiploidy is associated with an exceptionally poor prog-
otherwise typical lymphoblasts. Because of differences in monoclonal nosis. 88,89,92 Flow cytometric determination of cellular DNA content is
antibodies and immunophenotyping techniques, the frequencies of a useful adjunct to cytogenetic analysis because it is automated, rapid,
myeloid-associated antigen expression range from 5 to 30 percent in and inexpensive, and its measurements are not affected by the mitotic
childhood cases and from 10 to 50 percent in adult cases. 67,84 The pat- index of the cell population; results can be obtained in almost all cases.
tern of myeloid-associated antigen expression is correlated with certain Flow cytometric studies can sometimes identify a small but drug-resis-
genetic features of blast cells. CD15, CD33, and CD65 are expressed tant subpopulation of near-haploid cells that may be missed by standard
in ALL cases with a rearranged MLL gene, and CD13 and CD33 are cytogenetic analysis.
expressed in cases with the ETV6-RUNX1. There is a subset of cases Reciprocal chromosomal translocations in cases of B-cell and
84
that coexpress both lymphoid and myeloid markers but do not cluster T-cell ALL arise from errors in the normal recombination mecha-
with T-cell, B-cell precursor, or acute myeloid leukemia in gene-expression nisms that generate antigen receptor genes. Such rearrangements can
profiling. These cases may not respond to myeloid-directed therapy but fuse the promoter/enhancer element of the immunoglobulin heavy- or
attain remission with ALL-directed induction treatment. 85,86 The pres- light-chain gene or the T-cell antigen receptor β/γ or α/δ gene to sites
ence of myeloid-associated antigens lacks prognostic significance in adjacent to a variety of transcription factor genes. More often, genetic
contemporary treatment programs but can be useful in immunologic rearrangements result from the fusion of two genes encoding different
monitoring of patients for minimal residual leukemia. 67,83 transcription factors. These chimeric genes encode active kinases and
altered transcription factors that regulate genes involved in the differen-
Genetic Classification tiation, self-renewal, proliferation, and drug resistance of hematopoietic
ALL arises from a lymphoid progenitor cell that has sustained multiple stem cells.
specific genetic alterations that lead to malignant transformation and Specific cytogenetic findings are correlated with presenting clinical
proliferation. Thus, genetic classification of ALL yields more relevant features, blast-cell phenotypes, and clinical outcome (see Table 91–5).
biologic information than any other means. Approximately 75 per- However, there are now compelling reasons to focus on molecular
cent of adult and childhood cases can be readily classified into prog- genetic lesions. First, molecular analyses can identify important sub-
nostically or therapeutically relevant subgroups based on the modal microscopic genetic alterations not visible by standard karyotyping,
chromosome number (or DNA content estimated by flow cytometry), such as the ETV6-RUNX1 fusion, intrachromosomal amplification of
specific chromosomal rearrangements, and molecular genetic chan chromosome 21, deletions of tumor-suppressor genes, and mutations of
ges. 1,2,17–20,41,54–56,87–89 Table 91–5 summarizes the prominent clinical and protooncogenes. 1,2,87,93–95 Second, cases with clinically important genetic
biologic features of cases with the most common genetic abnormalities. rearrangements can be missed because of technical errors (e.g., karyo-
Two ploidy groups (hyperdiploidy >50 chromosomes and hypo- typing residual normal metaphase cells rather than leukemic metaphase
diploidy <44 chromosomes) have clinical relevance. Hyperdiploidy, cells). Hence, FISH and reverse transcriptase polymerase chain reaction
Kaushansky_chapter 91_p1505-1526.indd 1512 9/21/15 12:19 PM