Page 202 - Williams Hematology ( PDFDrive )
P. 202
176 Part IV: Molecular and Cellular Hematology Chapter 13: Cytogenetics and Genetic Abnormalities 177
A B
C D
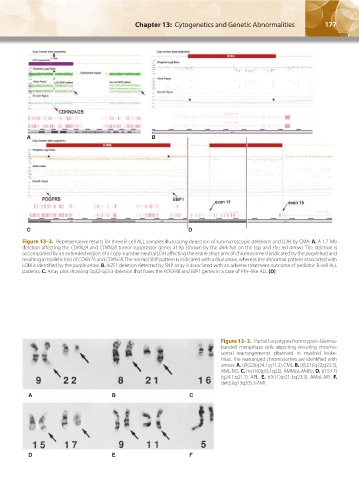
Figure 13–2. Representative results for three B-cell ALL samples illustrating detection of submicroscopic deletions and LOH by CMA. A. A 1.7-Mb
deletion affecting the CDKN2A and CDKN2B tumor suppressor genes at 9p (shown by the dark bar on the top and the red arrow). The deletion is
accompanied by an extended region of a copy-number neutral LOH affecting the entire short arm of chromosome 9 (indicated by the purple bar) and
resulting in biallelic loss of CDKN2A and CDKN2B. The normal SNP pattern is indicated with a blue arrow, whereas the abnormal pattern associated with
LOH is identified by the purple arrow. B. IKZF1 deletion detected by SNP array is associated with an adverse treatment outcome of pediatric B-cell ALL
patients. C. Array plot showing 5q32-q33.3 deletion that fuses the PDGFRB and EBF1 genes in a case of Ph+-like ALL (D).
Figure 13–3. Partial karyotypes from trypsin-Giemsa-
banded metaphase cells depicting recurring chromo-
somal rearrangements observed in myeloid leuke-
mias. The rearranged chromosomes are identified with
arrows. A. t(9;22)(q34.1;q11.2), CML. B. t(8;21)(q22;q22.3),
AML-M2. C. inv(16)(pl3.1q22), AMMoL-M4Eo. D. t(15;17)
(q24.1;q21.1), APL. E. t(9;11)(p21.3;q23.3), AMoL-M5. F.
del(5)(q13q33), t-AML.
A B C
D E F
Kaushansky_chapter 13_p0173-0190.indd 177 17/09/15 6:32 pm