Page 1087 - Clinical Immunology_ Principles and Practice ( PDFDrive )
P. 1087
ChaPTEr 78 Lymphoid Leukemias 1051
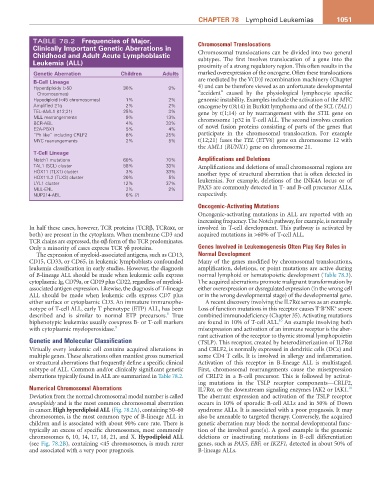
TABLE 78.2 Frequencies of Major, Chromosomal Translocations
Clinically important genetic aberrations in
Childhood and adult acute Lymphoblastic Chromosomal translocations can be divided into two general
subtypes. The first involves translocation of a gene into the
Leukemia (aLL) proximity of a strong regulatory region. This often results in the
genetic aberration Children adults marked overexpression of the oncogene. Often these translocations
B-Cell Lineage are mediated by the V(D)J recombination machinery (Chapter
Hyperdiploidy (>50 30% 9% 4) and can be therefore viewed as an unfortunate developmental
Chromosomes) “accident” caused by the physiological lymphocyte specific
Hypodiploid (<45 chromosomes) 1% 2% genomic instability. Examples include the activation of the MYC
Amplified 21q 2% 2% oncogene by t(8;14) in Burkitt lymphoma and of the SCL (TAL1)
TEL-AML1 (t12;21) 25% 3% gene by t(1;14) or by rearrangement with the STIL gene on
MLL rearrangements 9% 13% chromosome 1p32 in T-cell ALL. The second involves creation
BCR-ABL 4% 33%
E2A-PBX1 5% 4% of novel fusion proteins consisting of parts of the genes that
“Ph like” including CRLF2 8% 25% participate in the chromosomal translocation. For example
MYC rearrangements 2% 5% t(12;21) fuses the TEL (ETV6) gene on chromosome 12 with
the AML1 (RUNX1) gene on chromosome 21.
T-Cell Lineage
Notch1 mutations 60% 70% Amplifications and Deletions
TAL1 (SCL) cluster 58% 33% Amplifications and deletions of small chromosomal regions are
HOX11 (TLX1) cluster 3% 33% another type of structural aberration that is often detected in
HOX11L2 (TLX3) cluster 20% 5%
LYL1 cluster 12% 37% leukemias. For example, deletions of the INK4A locus or of
MLL-ENL 2% 2% PAX5 are commonly detected in T- and B-cell precursor ALLs,
NUP214-ABL 6% (?) respectively.
Oncogenic-Activating Mutations
Oncogenic-activating mutations in ALL are reported with an
increasing frequency. The Notch pathway, for example, is normally
In half these cases, however, TCR proteins (TCRβ, TCRαα, or involved in T-cell development. This pathway is activated by
both) are present in the cytoplasm. When membrane CD3 and acquired mutations in >60% of T-cell ALL.
TCR chains are expressed, the αβ form of the TCR predominates.
Only a minority of cases express TCR γδ proteins. Genes Involved in Leukemogenesis Often Play Key Roles in
The expression of myeloid-associated antigens, such as CD13, Normal Development
CD15, CD33, or CD65, in leukemic lymphoblasts confounded Many of the genes modified by chromosomal translocations,
leukemia classification in early studies. However, the diagnosis amplification, deletions, or point mutations are active during
of B-lineage ALL should be made when leukemic cells express normal lymphoid or hematopoietic development (Table 78.3).
cytoplasmic Ig, CD79a, or CD19 plus CD22, regardless of myeloid- The acquired aberrations promote malignant transformation by
associated antigen expression. Likewise, the diagnosis of T-lineage either overexpression or dysregulated expression (in the wrong cell
ALL should be made when leukemic cells express CD7 plus or in the wrong developmental stage) of the developmental gene.
either surface or cytoplasmic CD3. An immature immunophe- A recent discovery involving the IL7Rα serves as an example.
+
− +
notype of T-cell ALL, early T phenotype (ETP) ALL, has been Loss of function mutations in this receptor causes T B NK severe
8
described and is similar to normal ETP precursors. True combined immunodeficiency (Chapter 35). Activating mutations
10
biphenotypic leukemias usually coexpress B- or T-cell markers are found in 10% of T-cell ALL. An example involving both
with cytoplasmic myeloperoxidase. 9 misexpression and activation of an immune receptor is the aber-
rant activation of the receptor to thymic stromal lymphopoietin
Genetic and Molecular Classification (TSLP). This receptor, created by heterodimerization of IL7Rα
Virtually every leukemic cell contains acquired alterations in and CRLF2, is normally expressed in dendritic cells (DCs) and
multiple genes. These alterations often manifest gross numerical some CD4 T cells. It is involved in allergy and inflammation.
or structural aberrations that frequently define a specific clinical Activation of this receptor in B-lineage ALL is multistaged.
subtype of ALL. Common and/or clinically significant genetic First, chromosomal rearrangements cause the misexpression
aberrations typically found in ALL are summarized in Table 78.2. of CRLF2 in a B-cell precursor. This is followed by activat-
ing mutations in the TSLP receptor components—CRLF2,
Numerical Chromosomal Aberrations IL7Rα, or the downstream signaling enzymes JAK2 or JAK1.
10
Deviation from the normal chromosomal modal number is called The aberrant expression and activation of the TSLP receptor
aneuploidy and is the most common chromosomal aberration occurs in 10% of sporadic B-cell ALLs and in 50% of Down
in cancer. High hyperdiploid ALL (Fig. 78.2A), containing 50–60 syndrome ALLs. It is associated with a poor prognosis. It may
chromosomes, is the most common type of B-lineage ALL in also be amenable to targeted therapy. Conversely, the acquired
children and is associated with about 90% cure rate. There is genetic aberration may block the normal developmental func-
typically an excess of specific chromosomes, most commonly tion of the involved gene(s). A good example is the genomic
chromosomes 6, 10, 14, 17, 18, 21, and X. Hypodiploid ALL deletions or inactivating mutations in B-cell differentiation
(see Fig. 78.2B), containing <45 chromosomes, is much rarer genes, such as PAX5, EBF, or IKZF1, detected in about 50% of
and associated with a very poor prognosis. B-lineage ALLs.