Page 487 - Clinical Immunology_ Principles and Practice ( PDFDrive )
P. 487
468 ParT FOUr Immunological Deficiencies
Mutation on founder chromosome X
X X X X
Fragmentation of original
chromosome by recombination X X X
as population expands through
multiple generations
X X X
X
X X X X
X
X X X X
X
X X X X
X
X X X
X X X X
Mutation located within region of
linkage disequilibrium
A B
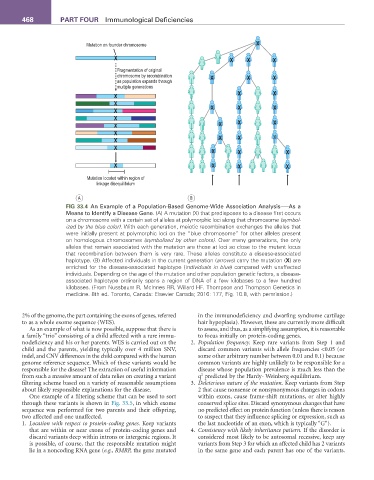
FIG 33.4 An Example of a Population-Based Genome-Wide Association Analysis–—As a
Means to Identify a Disease Gene. (A) A mutation (X) that predisposes to a disease first occurs
on a chromosome with a certain set of alleles at polymorphic loci along that chromosome (symbol-
ized by the blue color). With each generation, meiotic recombination exchanges the alleles that
were initially present at polymorphic loci on the “blue chromosome” for other alleles present
on homologous chromosomes (symbolized by other colors). Over many generations, the only
alleles that remain associated with the mutation are those at loci so close to the mutant locus
that recombination between them is very rare. These alleles constitute a disease-associated
haplotype. (B) Affected individuals in the current generation (arrows) carry the mutation (X) are
enriched for the disease-associated haplotype (individuals in blue) compared with unaffected
individuals. Depending on the age of the mutation and other population genetic factors, a disease-
associated haplotype ordinarily spans a region of DNA of a few kilobases to a few hundred
kilobases. (From Nussbaum R, McInnes RR, Willard HF. Thompson and Thompson Genetics in
medicine. 8th ed. Toronto, Canada: Elsevier Canada; 2016: 177, Fig. 10.8, with permission.)
2% of the genome, the part containing the exons of genes, referred in the immunodeficiency and dwarfing syndrome cartilage
to as a whole exome sequence (WES). hair hypoplasia). However, these are currently more difficult
As an example of what is now possible, suppose that there is to assess, and thus, as a simplifying assumption, it is reasonable
a family “trio” consisting of a child affected with a rare immu- to focus initially on protein-coding genes.
nodeficiency and his or her parents. WES is carried out on the 2. Population frequency. Keep rare variants from Step 1 and
child and the parents, yielding typically over 4 million SNV, discard common variants with allele frequencies <0.05 (or
indel, and CNV differences in the child compared with the human some other arbitrary number between 0.01 and 0.1) because
genome reference sequence. Which of these variants would be common variants are highly unlikely to be responsible for a
responsible for the disease? The extraction of useful information disease whose population prevalence is much less than the
2
from such a massive amount of data relies on creating a variant q predicted by the Hardy- Weinberg equilibrium.
filtering scheme based on a variety of reasonable assumptions 3. Deleterious nature of the mutation. Keep variants from Step
about likely responsible explanations for the disease. 2 that cause nonsense or nonsynonymous changes in codons
One example of a filtering scheme that can be used to sort within exons, cause frame-shift mutations, or alter highly
through these variants is shown in Fig. 33.5, in which exome conserved splice sites. Discard synonymous changes that have
sequence was performed for two parents and their offspring, no predicted effect on protein function (unless there is reason
two affected and one unaffected. to suspect that they influence splicing or expression, such as
1. Location with respect to protein-coding genes. Keep variants the last nucleotide of an exon, which is typically “G”).
that are within or near exons of protein-coding genes and 4. Consistency with likely inheritance pattern. If the disorder is
discard variants deep within introns or intergenic regions. It considered most likely to be autosomal recessive, keep any
is possible, of course, that the responsible mutation might variants from Step 3 for which an affected child has 2 variants
lie in a noncoding RNA gene (e.g., RMRP, the gene mutated in the same gene and each parent has one of the variants.