Page 488 - Clinical Immunology_ Principles and Practice ( PDFDrive )
P. 488
CHaPTEr 33 Human Genomics in Immunology 469
FILTERS or in an animal model with the disease? Finding mutations in
one of the candidate genes also in other patients would confirm
that this gene was the responsible gene trio under study. In some
Known 1° cases, one gene remaining on the list in Step 4 may become a
Recessive or
Sequence immuno- leading candidate because its involvement makes biological or
quality MAF≤0.05 de novo deficiency genetic sense or it is known to be mutated in other affected
pedigree
genes individuals. In other cases, however, the gene responsible may
turn out to be entirely unanticipated on biological grounds or
17613 14353 88 2
protein high 3901 recessive variants in may not be mutated in other affected individuals because of
altering quality rare or de novo one gene-> locus heterogeneity (i.e., mutations in other as yet undiscovered
variants variants variants variants ZAP-70 genes can cause a similar disease).
3260 10452 3813 86 Such variant assessments require extensive use of public
genomic databases and software tools. These include the human
genome reference sequence, databases of variants with their allelic
frequencies, software that assesses how deleterious an amino
acid substitution might be to gene function, collections of known
disease-causing mutations, and databases of functional networks
Set aside and biological pathways. The enormous expansion of this
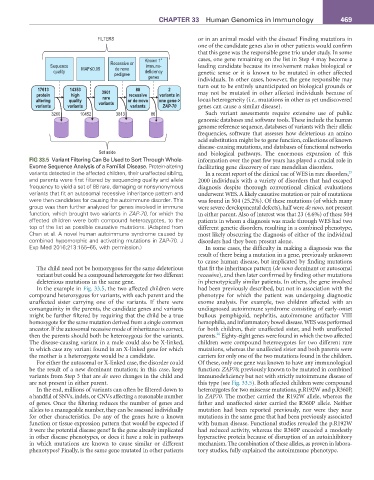
FIG 33.5 Variant Filtering Can Be Used to Sort Through Whole- information over the past few years has played a crucial role in
Exome Sequence Analysis of a Familial Disease. Protein-altering facilitating gene discovery of rare mendelian disorders.
variants detected in the affected children, their unaffected sibling, In a recent report of the clinical use of WES in rare disorders,
25
and parents were first filtered by sequencing quality and allele 2000 individuals with a variety of disorders that had escaped
frequency to yield a set of 88 rare, damaging or nonsynonymous diagnosis despite thorough conventional clinical evaluations
variants that fit an autosomal recessive inheritance pattern and underwent WES. A likely causative mutation or pair of mutations
were then candidates for causing the autoimmune disorder. This was found in 504 (25.2%). Of these mutations (of which many
group was then further analyzed for genes involved in immune were severe developmental defects), half were de novo, not present
function, which brought two variants in ZAP-70, for which the in either parent. Also of interest was that 23 (4.6%) of these 504
affected children were both compound heterozygotes, to the patients in whom a diagnosis was made through WES had two
top of the list as possible causative mutations. (Adapted from different genetic disorders, resulting in a combined phenotype,
Chan et al. A novel human autoimmune syndrome caused by most likely obscuring the diagnosis of either of the individual
combined hypomorphic and activating mutations in ZAP-70. J disorders had they been present alone.
Exp Med 2016;213:155–65, with permission.) In some cases, the difficulty in making a diagnosis was the
result of there being a mutation in a gene, previously unknown
to cause human disease, but implicated by finding mutations
The child need not be homozygous for the same deleterious that fit the inheritance pattern (de novo dominant or autosomal
variant but could be a compound heterozygote for two different recessive), and then later confirmed by finding other mutations
deleterious mutations in the same gene. in phenotypically similar patients. In others, the gene involved
In the example in Fig. 33.5, the two affected children were had been previously described, but not in association with the
compound heterozygous for variants, with each parent and the phenotype for which the patient was undergoing diagnostic
unaffected sister carrying one of the variants. If there were exome analysis. For example, two children affected with an
consanguinity in the parents, the candidate genes and variants undiagnosed autoimmune syndrome consisting of early-onset
might be further filtered by requiring that the child be a true bullous pemphigoid, nephritis, autoimmune antifactor VIII
homozygote for the same mutation derived from a single common hemophilia, and inflammatory bowel disease. WES was performed
ancestor. If the autosomal recessive mode of inheritance is correct, for both children, their unaffected sister, and both unaffected
26
then the parents should both be heterozygous for the variants. parents. Eighty-eight genes were found in which the two affected
The disease-causing variant in a male could also be X-linked, children were compound heterozygotes for two different rare
in which case any variant found in an X-linked gene for which mutations, whereas the unaffected sister and both parents were
the mother is a heterozygote would be a candidate. carriers for only one of the two mutations found in the children.
For either the autosomal or X-linked case, the disorder could Of these, only one gene was known to have any immunological
be the result of a new dominant mutation; in this case, keep function: ZAP70, previously known to be mutated in combined
variants from Step 3 that are de novo changes in the child and immunodeficiency but not with strictly autoimmune disease of
are not present in either parent. this type (see Fig. 33.5). Both affected children were compound
In the end, millions of variants can often be filtered down to heterozygotes for two missense mutations, p.R192W and p.R360P,
a handful of SNVs, indels, or CNVs affecting a reasonable number in ZAP70. The mother carried the R192W allele, whereas the
of genes. Once the filtering reduces the number of genes and father and unaffected sister carried the R360P allele. Neither
alleles to a manageable number, they can be assessed individually mutation had been reported previously, nor were they near
for other characteristics. Do any of the genes have a known mutations in the same gene that had been previously associated
function or tissue expression pattern that would be expected if with human disease. Functional studies revealed the p.R192W
it were the potential disease gene? Is the gene already implicated had reduced activity, whereas the R360P encoded a modestly
in other disease phenotypes, or does it have a role in pathways hyperactive protein because of disruption of an autoinhibitory
in which mutations are known to cause similar or different mechanism. The combination of these alleles, as proven in labora-
phenotypes? Finally, is the same gene mutated in other patients tory studies, fully explained the autoimmune phenotype.