Page 1400 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 1400
1246 Part VII Hematologic Malignancies
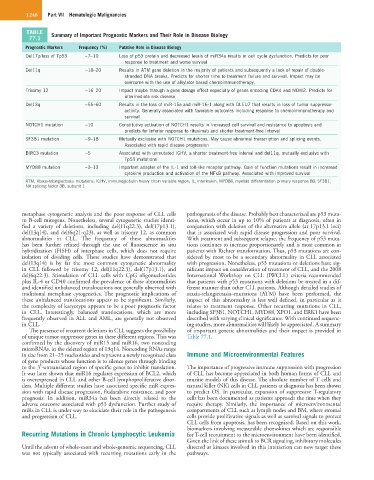
TABLE Summary of Important Prognostic Markers and Their Role in Disease Biology
77.1
Prognostic Markers Frequency (%) Putative Role in Disease Biology
Del17p/loss of Tp53 ~7–10 Loss of p53 protein and decreased levels of miR34a results in cell cycle dysfunction. Predicts for poor
response to treatment and worse survival
Del11q ~18–20 Results in ATM gene deletion in the majority of patients and subsequently a lack of repair of double-
stranded DNA breaks. Predicts for shorter time to treatment failure and survival. Impact may be
overcome with the use of alkylator based chemoimmunotherapy.
Trisomy 12 ~16–20 Impact maybe through a gene dosage effect especially of genes encoding CDK4 and MDM2. Predicts for
intermediate risk disease
Del13q ~55–60 Results in the loss of miR-15a and miR-16-1 along with DLEU7 that results in loss of tumor suppressor
activity. Generally associated with favorable outcomes including response to chemoimmunotherapy and
survival
NOTCH1 mutation ~10 Constitutive activation of NOTCH1 results in increased cell survival and resistance to apoptosis and
predicts for inferior response to rituximab and shorter treatment-free interval
SF3B1 mutation ~9–18 Mutually exclusive with NOTCH1 mutations. May cause abnormal transcription and splicing events.
Associated with rapid disease progression
BIRC3 mutation ~5 Associated with unmutated IGHV, a shorter treatment-free interval and del11q. mutually exclusive with
Tp53 mutations
MYD88 mutation ~3–13 Important adapter of the IL-1 and toll-like receptor pathway. Gain of function mutations result in increased
cytokine production and activation of the NFκB pathway. Associated with improved survival
ATM, Ataxia-telangiectasia–mutations; IGHV, immunoglobulin heavy chain variable region; IL, interleukin; MYD88, myeloid differentiation primary response 88; SF3B1,
NA splicing factor 3B, subunit 1
metaphase cytogenetic analysis and the poor response of CLL cells pathogenesis of the disease. Probably best characterized are p53 muta-
to B-cell mitogens. Nonetheless, several cytogenetic studies identi- tions, which occur in up to 10% of patients at diagnosis, often in
fied a variety of deletions, including del(11q22.3), del(17p13.1), conjunction with deletion of the alternative allele (at 17p13.1 loci)
del(13q14), and del(6q21-q23), as well as trisomy 12, as common that is associated with rapid disease progression and poor survival.
abnormalities in CLL. The frequency of these abnormalities With treatment and subsequent relapse, the frequency of p53 muta-
has been further refined through the use of fluorescence in situ tions continues to increase proportionately and is most common in
hybridization (FISH) of interphase cells, which does not require patients with Richter transformation. Thus, p53 mutations are con-
isolation of dividing cells. These studies have demonstrated that sidered by most to be a secondary abnormality in CLL associated
del(13q14) is by far the most common cytogenetic abnormality with progression. Nonetheless, p53 mutations or deletions have sig-
in CLL followed by trisomy 12, del(11q22.3), del(17p13.1), and nificant impact on consideration of treatment of CLL, and the 2008
del(6q22.3). Stimulation of CLL cells with CpG oligonucleotides International Workshop on CLL (IWCLL) criteria recommended
plus IL-4 or CD40 confirmed the prevalence of these abnormalities that patients with p53 mutations with deletions be treated in a dif-
and identified unbalanced translocations not generally observed with ferent manner than other CLL patients. Although detailed studies of
traditional metaphase cytogenetics. The prognostic implications of ataxia-telangiectasia–mutations (ATM) have been performed, the
these unbalanced translocations appear to be significant. Similarly, impact of this abnormality is less well defined, in particular as it
the complexity of karyotype appears to be a poor prognostic factor relates to treatment response. Other recurring mutations in CLL,
in CLL. Interestingly, balanced translocations, which are more including SF3B1, NOTCH1, MYD88, XPO1, and ERK1 have been
frequently observed in ALL and AML, are generally not observed described with varying clinical significance. With continued sequenc-
in CLL. ing studies, more abnormalities will likely be appreciated. A summary
The presence of recurrent deletions in CLL suggests the possibility of important genetic abnormalities and their impact is provided in
of unique tumor suppressor genes in these different regions. This was Table 77.1.
confirmed by the discovery of miR15 and miR16, two noncoding
microRNAs, in the deleted region of 13q14. Noncoding RNAs range
in size from 21–25 nucleotides and represent a newly recognized class Immune and Microenvironmental Features
of gene products whose function is to silence genes through binding
to the 3′-untranslated region of specific genes to inhibit translation. The importance of progressive immune suppression with progression
It was later shown that miR16 regulates expression of BCL2, which of CLL has become appreciated in both human forms of CLL and
is overexpressed in CLL and other B-cell lymphoproliferative disor- murine models of this disease. The absolute number of T cells and
ders. Multiple different studies have associated specific miR expres- natural killer (NK) cells in CLL patients at diagnosis has been shown
sion with rapid disease progression, fludarabine resistance, and poor to predict OS. In particular, expansion of suppressive T-regulatory
prognosis. In addition, miR34a has been directly related to the cells has been documented as patients approach the time when they
adverse outcome associated with p53 dysfunction. Further study of require therapy. Similarly, the importance of microenvironmental
miRs in CLL is under way to elucidate their role in the pathogenesis compartments of CLL such as lymph nodes and BM, where stromal
and progression of CLL. cells provide proliferative signals as well as survival signals to protect
CLL cells from apoptosis, has been recognized. Based on this work,
biomarkers involving measurable chemokines which are responsible
Recurring Mutations in Chronic Lymphocytic Leukemia for T-cell recruitment to the microenvironment have been identified.
Given the link of these stimuli to BCR signaling, inhibitory molecules
Until the advent of whole-exon and whole-genomic sequencing, CLL directed at kinases involved in this interaction can now target these
was not typically associated with recurring mutations early in the pathways.