Page 1396 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 1396
1242 Part VII Hematologic Malignancies
GCB
ABC
MCL SLL DLBCL DLBCL
BANK
Arachidonate 5-LO
MARCKS
CD1D Fold
MMP-11 Relative
MD-1 Expression
4x
CD20 2x
IRTA4
CALL
G gamma 11 1x
TLR10
NKG2D 0.5x
CD24
EphB6
CEA-related
adhesion 1 0.25x
MASL1
hCAP1a
IRTA5
GPCR W
Wnt-3
IGF binding prot. 3
Lymphoma Biopsies
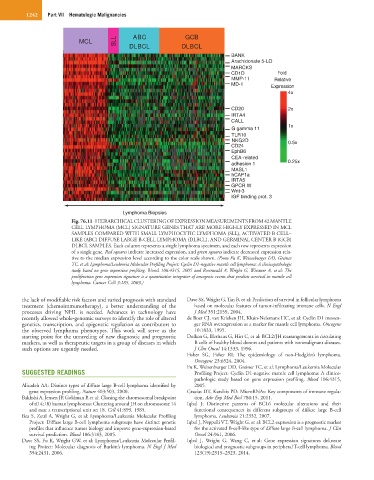
Fig. 76.11 HIERARCHICAL CLUSTERING OF EXPRESSION MEASUREMENTS FROM 42 MANTLE
CELL LYMPHOMA (MCL) SIGNATURE GENES THAT ARE MORE HIGHLY EXPRESSED IN MCL
SAMPLES COMPARED WITH SMALL LYMPHOCYTIC LYMPHOMA (SLL), ACTIVATED B CELL–
LIKE (ABC) DIFFUSE LARGE B-CELL LYMPHOMA (DLBCL), AND GERMINAL CENTER B (GCB)
DLBCL SAMPLES. Each column represents a single lymphoma specimen, and each row represents expression
of a single gene. Red squares indicate increased expression, and green squares indicate decreased expression rela-
tive to the median expression level according to the color scale shown. (From Fu K, Weisenburger DD, Greiner
TC, et al: Lymphoma/Leukemia Molecular Profiling Project: Cyclin D1-negative mantle cell lymphoma: A clinicopathologic
study based on gene expression profiling. Blood 106:4315, 2005 and Rosenwald A, Wright G, Wiestner A, et al: The
proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell
lymphoma. Cancer Cell 3:185, 2003.)
the lack of modifiable risk factors and varied prognosis with standard Dave SS, Wright G, Tan B, et al: Prediction of survival in follicular lymphoma
treatment (chemoimmunotherapy), a better understanding of the based on molecular features of tumor-infiltrating immune cells. N Engl
processes driving NHL is needed. Advances in technology have J Med 351:2159, 2004.
recently allowed whole-genomic surveys to identify the role of altered de Boer CJ, van Krieken JH, Kluin-Nelemans HC, et al: Cyclin D1 messen-
genetics, transcription, and epigenetic regulation as contributors to ger RNA overexpression as a marker for mantle cell lymphoma. Oncogene
the observed lymphoma phenotypes. This work will serve as the 10:1833, 1995.
starting point for the unraveling of new diagnostic and prognostic Dolken G, Illerhaus G, Hirt C, et al: BCL2/JH rearrangements in circulating
markers, as well as therapeutic targets in a group of diseases in which B cells of healthy blood donors and patients with nonmalignant diseases.
such options are urgently needed. J Clin Oncol 14:1333, 1996.
Fisher SG, Fisher RI: The epidemiology of non-Hodgkin’s lymphoma.
Oncogene 23:6524, 2004.
Fu K, Weisenburger DD, Greiner TC, et al: Lymphoma/Leukemia Molecular
SUGGESTED READINGS Profiling Project: Cyclin D1-negative mantle cell lymphoma: A clinico-
pathologic study based on gene expression profiling. Blood 106:4315,
Alizadeh AA: Distinct types of diffuse large B-cell lymphoma identified by 2005.
gene expression profiling. Nature 403:503, 2000. Gracias DT, Katsikis PD: MicroRNAs: Key components of immune regula-
Bakhshi A, Jensen JP, Goldman P, et al: Cloning the chromosomal breakpoint tion. Adv Exp Med Biol 780:15, 2011.
of t(14;18) human lymphomas: Clustering around JH on chromosome 14 Iqbal J: Distinctive patterns of BCL6 molecular alterations and their
and near a transcriptional unit on 18. Cell 41:899, 1985. functional consequences in different subgroups of diffuse large B-cell
Bea S, Zettl A, Wright G, et al: Lymphoma/Leukemia Molecular Profiling lymphoma. Leukemia 21:2332, 2007.
Project: Diffuse large B-cell lymphoma subgroups have distinct genetic Iqbal J, Neppalli VT, Wright G, et al: BCL2 expression is a prognostic marker
profiles that influence tumor biology and improve gene-expression-based for the activated B-cell-like type of diffuse large B-cell lymphoma. J Clin
survival prediction. Blood 106:3183, 2005. Oncol 24:961, 2006.
Dave SS, Fu K, Wright GW, et al: Lymphoma/Leukemia Molecular Profil- Iqbal J, Wright G, Wang C, et al: Gene expression signatures delineate
ing Project: Molecular diagnosis of Burkitt’s lymphoma. N Engl J Med biological and prognostic subgroups in peripheral T-cell lymphoma. Blood
354:2431, 2006. 123(19):2915–2923, 2014.