Page 331 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 331
Chapter 24 Complement and Immunoglobulin Biology Leading to Clinical Translation 273
monomer that contains four polypeptide chains: two identical heavy immunoglobulin, also interacts with protein A, an immune evasion
(H) chains and two identical light (L) chains covalently linked by molecule on the cell walls of S. aureus. When bound to protein A,
139
disulfide bonds (Fig. 24.7). The x-ray crystallographic structure of the binding of IgG to host effector molecules such as C1q is sterically
a monomeric immunoglobulin, specifically a mouse IgG2a monoclo- interfered with.
nal antibody (mAb), is shown depicted in both ribbon and space- The chain structure of immunoglobulins explains neither anti-
140
filling models in Fig. 24.8. Depending on the angle between the body structural diversity nor antibody binding to antigen. The dis-
constituent Fab (fragment antigen-binding) monomers, an immuno- covery of variable and constant regions of amino acid sequence
globulin monomer consists of a Y- or T-like structure. The size of the formed the basis for understanding both phenomena. Thus in the L
Fab arms is 80 × 50 × 40 Å, and the size of base, called the Fc chain, the 100 or so amino acids in the amino-terminal half of the
(fragment crystallizable) region, is approximately 70 × 45 × 40 Å protein (variable region [V L ]) vary among antibody molecules, but in
according to the x-ray structure models. The Ig molecule exhibits the second half (constant region [C L ]), there is virtual complete
considerable flexibility. In electron microscopic, low-angle x-ray scat-
tering, transient electric birefringence, and resonance energy transfer
studies, the angle between the Fab domains has been observed to vary V L
from 0 to 180 degrees. All antibodies have two identical combining Fab C L Fab
sites for each antigen located at the ends of the Fab domains.
Fab and Fc represent functional domains in immunoglobulins.
They were discovered by performing limited proteolytic digestion of
the molecule. Both the H and L chains contribute amino acids that
constitute the antigen-binding site in Fab. The monovalent Fab frag- Hinge
ment will bind to, but will not precipitate, multivalent antigens, in V H C H 1
contrast to native IgG. A fragment can be prepared, called F(ab′) 2,
that is devoid of Fc but still precipitates antigen. This form of
immunoglobulin consists of two Fabs disulfide bonded at a part of C H 2
the molecule called the hinge region. The hinge region is the part
of the Ig molecule that is responsible for the molecular flexibility
exhibited by all immunoglobulins. The other major function of Fc
immunoglobulins, binding to specific receptors on cells and certain C H 3
effector proteins such as C1q, is associated with binding site(s) also
found in Fc. The Fc region of IgG, one of the classes of
A
H H
NH 2 NH 2
L CDR 1 L VL CL
V H VL
CDR 2
CDR 1 CH1 CL
CDR 3 D VH Hinge
CDR 2 J H V L region CH1
VH
CH 1 CH 1
CDR 3
CL CL J L
CH2
S-S SS S-S CH2
SS
CH 2 CH 2
CH3
B CH3
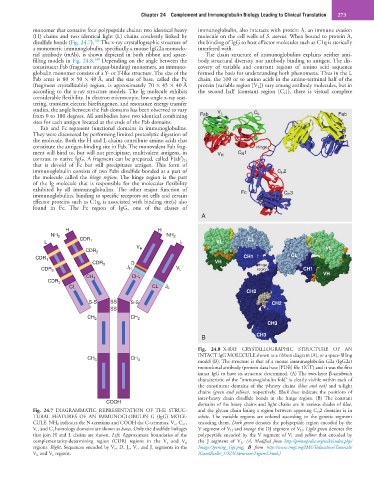
Fig. 24.8 X-RAY CRYSTALLOGRAPHIC STRUCTURE OF AN
INTACT IgG MOLECULE shown as a ribbon diagram (A), or a space-filling
CH 3 CH 3 model (B). The structure is that of a mouse immunoglobulin G2a (IgG2a)
monoclonal antibody (protein data base [PDB] file 1IGT) and it was the first
intact IgG to have its structure determined. (A) The two-layer β-sandwich
characteristic of the “immunoglobulin fold” is clearly visible within each of
the constituent domains of the γ-heavy chains (blue and red) and κ-light
chains (green and yellow), respectively. Black lines indicate the positions of
inter-heavy chain disulfide bonds in the hinge region. (B) The constant
COOH
domains of the heavy chains and light chains are in various shades of blue,
Fig. 24.7 DIAGRAMMATIC REPRESENTATION OF THE STRUC- and the glycan chain lining a region between apposing C H 2 domains is in
TURAL FEATURES OF AN IMMUNOGLOBULIN G (IgG) MOLE- white. The variable regions are colored according to the genetic segment
CULE. NH 2 indicates the N-terminus and COOH the C-terminus. V h , C h1 , encoding them. Dark green denotes the polypeptide region encoded by the
V l , and C l homology domains are shown as boxes. Only the disulfide linkages V segment of V H and orange the DJ segment of V H . Light green denotes the
that join H and L chains are shown. Left, Approximate boundaries of the polypeptide encoded by the V segment of V L and yellow that encoded by
complementarity-determining region (CDR) regions in the V l and V h the J segment of V L. (A, Modified from http://proteopedia.org/wiki/index.php/
regions. Right, Sequences encoded by V h , D, J h , V l, and J l segments in the Image:Opening_1igt.png; B from http://www.imgt.org/IMGTeducation/Tutorials/
V h and V l regions. IGandBcells/_UK/3Dstructure/Figure2.html.)