Page 928 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 928
Chapter 56 Conventional and Molecular Cytogenomic Basis of Hematologic Malignancies 811
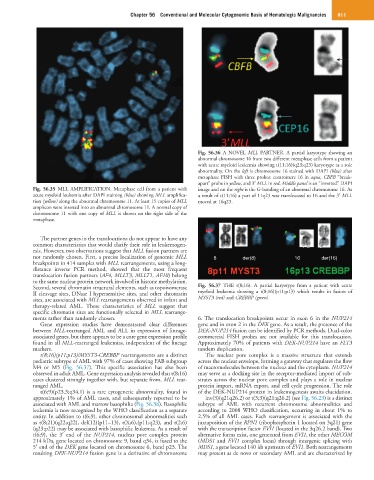
Fig. 56.36 A NOVEL MLL PARTNER. A partial karyotype showing an
abnormal chromosome 16 from two different metaphase cells from a patient
with acute myeloid leukemia showing t(11;16)(q23;q23) karyotype as a sole
abnormality. On the left is chromosome 16 stained with DAPI (blue) after
metaphase FISH with three probes: centromere 16 in aqua, CBFB “break-
apart” probe in yellow, and 3′ MLL in red. Middle panel is an “inverted” DAPI
Fig. 56.35 MLL AMPLIFICATION. Metaphase cell from a patient with image and on the right is the G-banding of an abnormal chromosome 16. As
acute myeloid leukemia after DAPI staining (blue) showing MLL amplifica- a result of t(11;16) a part of 11q23 was translocated to 16 and the 3′ MLL
tion (yellow) along the abnormal chromosome 11. At least 15 copies of MLL moved at 16q23.
amplicon were inserted into an abnormal chromosome 11. A normal copy of
chromosome 11 with one copy of MLL is shown on the right side of the
metaphase.
The partner genes in the translocations do not appear to have any
common characteristics that would clarify their role in leukemogen-
esis. However, two observations suggest that MLL fusion partners are
not randomly chosen. First, a precise localization of genomic MLL 8 der(8) 16 der(16)
breakpoints in 414 samples with MLL rearrangements, using a long-
distance inverse PCR method, showed that the most frequent
translocation fusion partners (AF4, MLLT3, MLLT1, AF10) belong
to the same nuclear protein network involved in histone methylation.
Second, several chromatin structural elements, such as topoisomerase Fig. 56.37 THE t(8;16). A partial karyotype from a patient with acute
II cleavage sites, DNase I hypersensitive sites, and other chromatin myeloid leukemia showing a t(8;16)(p11;p13) which results in fusion of
sites, are associated with MLL rearrangements observed in infant and MYST3 (red) and CREBBP (green).
therapy-related AML. These characteristics of MLL suggest that
specific chromatin sites are functionally selected in MLL rearrange-
ments rather than randomly chosen. 6. The translocation breakpoints occur in exon 6 in the NUP214
Gene expression studies have demonstrated clear differences gene and in exon 2 in the DEK gene. As a result, the presence of the
between MLL-rearranged AML and ALL in expression of lineage- DEK-NUP214 fusion can be identified by PCR methods. Dual-color
associated genes, but there appears to be a core gene expression profile commercial FISH probes are not available for this translocation.
found in all MLL-rearranged leukemias, independent of the lineage Approximately 70% of patients with DEK-NUP214 have an FLT3
markers. tandem duplication.
t(8;16)(p11;p13)/MYST3-CREBBP rearrangements are a distinct The nuclear pore complex is a massive structure that extends
pediatric subtype of AML with 97% of cases showing FAB subgroup across the nuclear envelope, forming a gateway that regulates the flow
M4 or M5 (Fig. 56.37). This specific association has also been of macromolecules between the nucleus and the cytoplasm. NUP214
observed in adult AML. Gene expression analysis revealed that t(8;16) may serve as a docking site in the receptor-mediated import of sub-
cases clustered strongly together with, but separate from, MLL rear- strates across the nuclear pore complex and plays a role in nuclear
ranged AML. protein import, mRNA export, and cell cycle progression. The role
t(6;9)(p23.3;q34.1) is a rare cytogenetic abnormality, found in of the DEK-NUP214 protein in leukemogenesis awaits elucidation.
approximately 1% of AML cases, and subsequently reported to be inv(3)(q21q26.2) or t(3;3)(q21;q26.2) (see Fig. 56.23) is a distinct
associated with AML and marrow basophilia (Fig. 56.38). Basophilic subtype of AML with recurrent chromosome abnormalities and
leukemia is now recognized by the WHO classification as a separate according to 2008 WHO classification, occurring in about 1% to
entity. In addition to t(6;9), other chromosomal abnormalities such 2.5% of all AML cases. Each rearrangement is associated with the
as t(8;21)(q22;q22), del(12)(p11–13), t(X;6),(p11;q23), and t(2;6) juxtaposition of the RPN1 (ribophosphorin 1 located on 3q21) gene
(q23;p22) may be associated with basophilic leukemia. As a result of with the transcription factor EVI1 (located in the 3q26.2 band). Two
t(6;9), the 3′ end of the NUP214, nuclear pore complex protein alternative forms exist, one generated from EVI1, the other MECOM
214 kDa, gene located on chromosome 9, band q34, is fused to the (MDS1 and EVI1 complex locus) through intergenic splicing with
5′ end of the DEK gene located on chromosome 6, band p23. The MDS1, a gene located 140 kb upstream of EVI1. Both rearrangements
resulting DEK-NUP214 fusion gene is a derivative of chromosome may present as de novo or secondary AML and are characterized by