Page 923 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 923
806 Part VII Hematologic Malignancies
A B C D
E
I
F H
G
J K
L
M
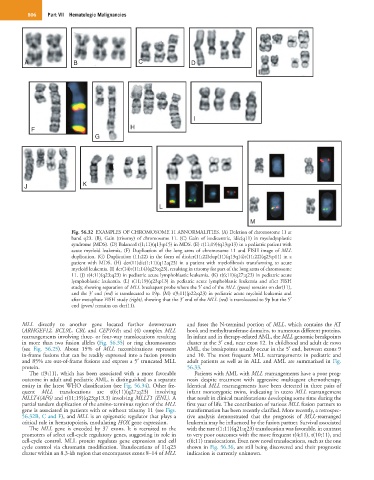
Fig. 56.32 EXAMPLES OF CHROMOSOME 11 ABNORMALITIES. (A) Deletion of chromosome 11 at
band q23. (B), Gain (trisomy) of chromosome 11. (C) Gain of isodicentric, idic(q11) in myelodysplastic
syndrome (MDS). (D) Balanced t(1;11)(q13;p15) in MDS. (E) t(11;19)(q13;p13) in a pediatric patient with
acute myeloid leukemia. (F) Duplication of the long arms of chromosome 11 and FISH image of MLL
duplication. (G) Duplication (11;22) in the form of dicder(11;22)dup(11)(q13q14)t(11;22)(q23;p11) in a
patient with MDS. (H) der(11)dic(1;11)(q12;q23) in a patient with myelofibrosis transforming to acute
myeloid leukemia. (I) der(14)t(11;14)(q23;q23), resulting in trisomy for part of the long arms of chromosome
11. (J) t(4;11)(q23;q23) in pediatric acute lymphoblastic leukemia. (K) t(6;11)(q27;q23) in pediatric acute
lymphoblastic leukemia. (L) t(11;19)(q23;p13) in pediatric acute lymphoblastic leukemia and after FISH
study, showing separation of MLL breakapart probe where the 5′ end of the MLL (green) remains on der(11),
and the 3′ end (red) is translocated to 19p. (M) t(9;11)(p22;q23) in pediatric acute myeloid leukemia and
after metaphase FISH study (right), showing that the 3′ end of the MLL (red) is translocated to 9p but the 5′
end (green) remains on der(11).
MLL directly to another gene located further downstream and fuses the N-terminal portion of MLL, which contains the AT
(ARHGEF12, BCL9L, CBL and CEP164); and (4) complex MLL hook and methyltransferase domains, to numerous different proteins.
rearrangements involving three- or four-way translocations resulting In infant and in therapy-related AML, the MLL genomic breakpoints
in more than two fusion alleles (Fig. 56.35) or ring chromosomes cluster at the 3′ end, near exon 12. In childhood and adult de novo
(see Fig. 56.25). About 15% of MLL recombinations represent AML, the breakpoints usually occur in the 5′ end, between exons 9
in-frame fusions that can be readily expressed into a fusion protein and 10. The most frequent MLL rearrangements in pediatric and
and 85% are out-of-frame fusions and express a 5′ truncated MLL adult patients as well as in ALL and AML are summarized in Fig.
protein. 56.33.
The t(9;11), which has been associated with a more favorable Patients with AML with MLL rearrangements have a poor prog-
outcome in adult and pediatric AML, is distinguished as a separate nosis despite treatment with aggressive multiagent chemotherapy.
entity in the latest WHO classification (see Fig. 56.34). Other fre- Identical MLL rearrangements have been detected in three pairs of
quent MLL translocations are t(6;11)(q27;q23) involving infant monozygotic twins, indicating in utero MLL rearrangement
MLLT4(AF6) and t(11;19)(q23;p13.3) involving MLLT1 (ENL). A that result in clinical manifestations developing some time during the
partial tandem duplication of the amino-terminus region of the MLL first year of life. The contribution of various MLL fusion partners to
gene is associated in patients with or without trisomy 11 (see Figs. transformation has been recently clarified. More recently, a retrospec-
56.32B, C and F), and MLL is an epigenetic regulator that plays a tive analysis demonstrated that the prognosis of MLL-rearranged
critical role in hematopoiesis, modulating HOX gene expression. leukemia may be influenced by the fusion partner. Survival associated
The MLL gene is encoded by 37 exons. It is recruited to the with the rare t(1;11)(q21;q23) translocation was favorable, in contrast
promoters of select cell-cycle regulatory genes, suggesting its role in to very poor outcomes with the more frequent t(4;11), t(10;11), and
cell-cycle control. MLL protein regulates gene expression and cell t(6;11) translocations. Even now novel translocations, such as the one
cycle control via chromatin modification. Translocations of 11q23 shown in Fig. 56.36, are still being discovered and their prognostic
cluster within an 8.3-kb region that encompasses exons 8–14 of MLL indication is currently unknown.