Page 932 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 932
Chapter 56 Conventional and Molecular Cytogenomic Basis of Hematologic Malignancies 815
BCR-ABL
(ASXL1)
(WT1)
PV CML
ET TET2 CSF3R
DNMT3A aCML SETBP1
JAK2 MPL
CALR
Spliceosome
ASXL1 TET2
DNMT3A ASXL1
RUNX1 MDS IMF CMML MLL-PTD
TET2 NRAS
FLT3
PML-RARA
CBFB-MYH11 TET2
RUNX1-RUNX1T1 Normal AML SM KIT
DNMT3A
NPM1
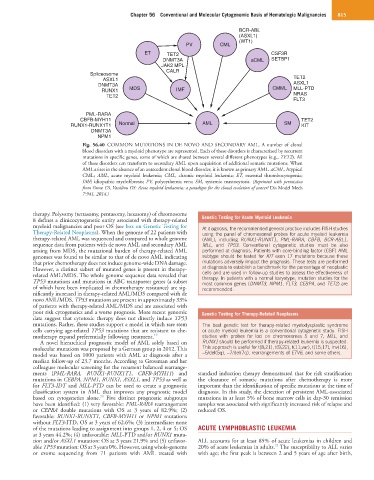
Fig. 56.40 COMMON MUTATIONS IN DE NOVO AND SECONDARY AML. A number of clonal
blood disorders with a myeloid phenotype are represented. Each of these disorders is characterized by recurrent
mutations in specific genes, some of which are shared between several different phenotypes (e.g., TET2). All
of these disorders can transform to secondary AML upon acquisition of additional somatic mutations. When
AML arises in the absence of an antecedent clonal blood disorder, it is known as primary AML. aCML, Atypical
CML; AML, acute myeloid leukemia; CML, chronic myeloid leukemia; ET, essential thrombocytopenia;
IMF, idiopathic myelofibrosis; PV, polycythemia vera; SM, systemic mastocytosis. (Reprinted with permission
from Grove CS, Vassiliou GS: Acute myeloid leukaemia: a paradigm for the clonal evolution of cancer? Dis Model Mech
7:941, 2014.)
therapy. Polysomy (tetrasomy, pentasomy, hexasomy) of chromosome
8 defines a clinicocytogenetic entity associated with therapy-related Genetic Testing for Acute Myeloid Leukemia
myeloid malignancies and poor OS (see box on Genetic Testing for At diagnosis, the recommended general practice includes FISH studies
Therapy-Related Neoplasms). When the genome of 22 patients with using the panel of chromosomal probes for acute myeloid leukemia
therapy-related AML was sequenced and compared to whole genome (AML), including RUNX1-RUNXT1, PML-RARA, CBFB, BCR-ABL1,
sequence data from patients with de novo AML and secondary AML MLL, and TP53. Conventional cytogenetic studies must be also
arising from MDS, the mutational burden of therapy-related AML performed at diagnosis. Patients with core-binding factor (CBF) AML
genomes was found to be similar to that of de novo AML indicating subtype should be tested for KIT exon 17 mutations because these
that prior chemotherapy does not induce genome-wide DNA damage. mutations adversely impact the prognosis. These tests are performed
However, a distinct subset of mutated genes is present in therapy- at diagnosis to establish a benchmark for the percentage of neoplastic
related AML/MDS. The whole genome sequence data revealed that cells and are used in follow-up studies to assess the effectiveness of
therapy. In patients with a normal karyotype, mutation studies for the
TP53 mutations and mutations in ABC transporter genes (a subset most common genes (DNMT3, NPM1, FLT3, CEBPA, and TET2) are
of which have been implicated in chemotherapy resistance) are sig- recommended.
nificantly increased in therapy-related AML/MDS compared with de
novo AML/MDS. TP53 mutations are present in approximately 33%
of patients with therapy-related AML/MDS and are associated with
poor risk cytogenetics and a worse prognosis. More recent genomic Genetic Testing for Therapy-Related Neoplasms
data suggest that cytotoxic therapy does not directly induce TP53
mutations. Rather, these studies support a model in which rare stem The best genetic test for therapy-related myelodysplastic syndrome
cells carrying age-related TP53 mutations that are resistant to che- or acute myeloid leukemia is a conventional cytogenetic study. FISH
motherapy expand preferentially following treatment. 19 studies with probes for loci on chromosomes 5 and 7, MLL, and
A novel hierarchical prognostic model of AML solely based on RUNX1 should be performed if therapy-related leukemia is suspected.
molecular mutations was proposed by a German group in 2012. This This approach is useful for t(8;21), t(9;22), t(11;var), t(15;17), inv(16),
model was based on 1000 patients with AML at diagnosis after a −5/del(5q), −7/del(7q), rearrangements of ETV6, and some others.
median follow-up of 23.7 months. According to Grossman and her
colleagues molecular screening for the recurrent balanced rearrange-
ments (PML-RARA, RUNX1-RUNX1T1, CBFB-MYH11) and standard induction therapy demonstrated that for risk stratification
mutations in CEBPA, NPM1, RUNX1, ASXL1, and TP53 as well as the clearance of somatic mutations after chemotherapy is more
for FLT3-IDT and MLL-PTD can be used to create a prognostic important than the identification of specific mutations at the time of
classification system in AML that improves any prognostic model diagnosis. In this study, the detection of persistent AML-associated
20
based on cytogenetics alone. Five distinct prognostic subgroups mutations in at least 5% of bone marrow cells in day-30 remission
have been identified: (1) very favorable: PML-RARA rearrangement samples was associated with significantly increased risk of relapse and
or CEPBA double mutations with OS at 3 years of 82.9%; (2) reduced OS.
favorable: RUNX1-RUNXT1, CBFB-MYH11 or NPM1 mutations
without FLT3-ITD, OS at 3 years of 62.6%; (3) intermediate: none
of the mutations leading to assignment into groups 1, 2, 4 or 5; OS ACUTE LYMPHOBLASTIC LEUKEMIA
at 3 years 44.2%; (4) unfavorable: MLL-PTD and/or RUNX1 muta-
tion and/or ASXL1 mutation: OS at 3 years 21.9% and (5) unfavor- ALL accounts for at least 85% of acute leukemias in children and
21
able TP53 mutation: OS at 3 years 0%. However, using whole-genome 20% of acute leukemias in adults. The susceptibility to ALL varies
or exome sequencing from 71 patients with AML treated with with age; the first peak is between 2 and 5 years of age after birth,