Page 1761 - Williams Hematology ( PDFDrive )
P. 1761
1736 Part XI: Malignant Lymphoid Diseases Chapter 107: Myeloma 1737
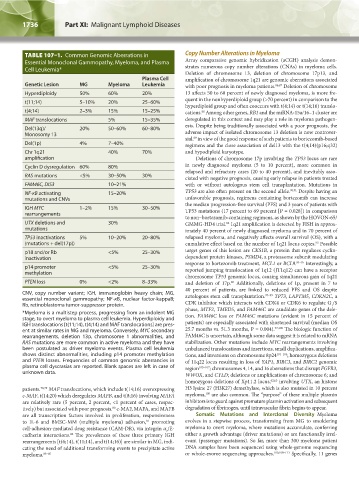
TABLE 107–1. Common Genomic Aberrations in Copy Number Alterations in Myeloma
Essential Monoclonal Gammopathy, Myeloma, and Plasma Array comparative genomic hybridization (aCGH) analysis demon-
strates numerous copy number alterations (CNAs) in myeloma cells.
Cell Leukemia*
Deletion of chromosome 13, deletion of chromosome 17p13, and
Plasma Cell amplification of chromosome 1q21 are genomic aberrations associated
Genetic Lesion MG Myeloma Leukemia with poor prognosis in myeloma patients. 86,87 Deletion of chromosome
Hyperdiploidy 50% 60% 20% 13 affects 50 to 60 percent of newly diagnosed myeloma, is more fre-
quent in the nonhyperdiploid group (>70 percent) in comparison to the
t(11;14) 5–10% 20% 25–60%
hyperdiploid group and often cooccurs with t(4;14) or t(14;16) translo-
t(4;14) 2–3% 15% 15–25% cations. Among other genes, RB1 and the miRNA-15a/16–1 cluster are
88
MAF translocations 5% 15–35% deregulated in this context and may play a role in myeloma pathogen-
esis. Despite being traditionally associated with a poor prognosis, the
Del(13q)/ 20% 50–60% 60–80%
Monosomy 13 adverse impact of isolated chromosome 13 deletion is now controver-
sial, in view of the good response of such patients to bortezomib-based
89
Del(1p) 4% 7–40% regimens and the close association of del13 with the t(4;14)(p16;q32)
Chr 1q21 40% 70% and hypodiploid karyotype.
amplification Deletions of chromosome 17p involving the TP53 locus are rare
Cyclin D dysregulation 60% 80% in newly diagnosed myeloma (5 to 10 percent), more common in
relapsed and refractory cases (20 to 40 percent), and inevitably asso-
RAS mutations <5% 30–50% 30% ciated with negative prognosis, causing early relapse in patients treated
FAM46C, DIS3 10–21% with or without autologous stem cell transplantation. Mutations in
NF-κB activating 15–20% TP53 are also often present on the second allele. 90,91 Despite having an
mutations and CNVs unfavorable prognosis, regimens containing bortezomib can increase
the median progression-free survival (PFS) and 3 years of patients with
IGH MYC 1–2% 15% 30–50% TP53 mutations (17 percent to 69 percent [P = 0.028]) in comparison
rearrangements
to non–bortezomib-containing regimens, as shown by the HOVON-65/
UTX deletions and 30% GMMG-HD4 trial. 1q21 amplification is detected by FISH in approx-
92
mutations imately 40 percent of newly diagnosed myeloma and in 70 percent of
TP53 inactivations 5% 10–20% 20–80% relapsed myeloma, and negatively affects overall survival (OS), with a
(mutations + del(17p)) cumulative effect based on the number of 1q21 locus copies. Possible
93
p18 and/or Rb <5% 25–30% target genes of this lesion are CKS1B, a protein that regulates cyclin-
inactivation dependent protein kinases, PSMD4, a proteasome subunit modulating
response to bortezomib treatment, MCL1 or BCL9. 93–95 Interestingly, a
p14 promoter <5% 25–30% reported jumping translocation of 1q12 (JT1q12) can have a receptor
methylation
chromosome TP53 genomic locus, causing simultaneous gain of 1q21
PTEN loss 0% <2% 8–33% and deletion of 17p. Additionally, deletions of 1p, present in 7 to
96
40 percent of patients, are linked to reduced PFS and OS despite
CNV, copy number variant; IGH, immunoglobin heavy chain; MG, autologous stem cell transplantation. 97–99 TP73, LAPTM5, CDKN2C, a
essential monoclonal gammopathy; NF-κB, nuclear factor-kappaB;
Rb, retinoblastoma tumor-suppressor protein. CDK inhibitor which interacts with CDK4 or CDK6 to regulate G /S
1
*Myeloma is a multistep process, progressing from an indolent MG phase, MTF2, TMED5, and FAM46C are candidate genes of the dele-
tion. FAM46C loss or FAM46C mutations (evident in 15 percent of
stage, to overt myeloma to plasma cell leukemia. Hyperdiploidy and
IGH translocations [t(11;14), t(4;14) and MAF translocations] are pres- patients) are especially associated with shortened survival (median OS
ent at similar rates in MG and myeloma. Conversely, MYC secondary 25.7 months vs. 51.3 months, P = 0.004). 97,100 The biologic function of
rearrangements, deletion 13p, chromosome 1 abnormalities, and FAM46C is uncertain, although some data suggest it is related to mRNA
RAS mutations are more common in active myeloma and they have stabilization. Other mutations include MYC rearrangements involving
been postulated as driver myeloma events. Plasma cell leukemia unbalanced translocations and insertions, small duplications, amplifica-
shows distinct abnormalities, including p14 promoter methylation tions, and inversions on chromosome 8p24 101–104 ; homozygous deletions
and PTEN losses. Frequencies of common genomic aberrancies in of 11q22 locus resulting in loss of YAP1, BIRC3, and BIRC2 genomic
plasma cell dyscrasias are reported. Blank spaces are left in case of region 105–107 ; chromosomes 4, 14, and 16 aberrations that disrupt FGFR3,
unknown data. WWOX, and CYLD; deletions or amplifications of chromosome 6; and
homozygous deletions of Xp11.2 locus, 62,63 involving UTX, an histone
patients. 78,79 MAF translocations, which include t(14;16) overexpressing H3 lysine 27 (H3K27) demethylase, which is also mutated in 10 percent
100
c-MAF, t(14;20) which deregulates MAFB, and t(8;16) involving MAFA myeloma, are also common. The “purpose” of these multiple plasmin
are relatively rare (5 percent, 2 percent, <1 percent of cases, respec- inhibitors is to guard against premature plasmin activation and subsequent
tively) but associated with poor prognosis. c-MAF, MAFA, and MAFB degradation of fibrinogen, until intravascular fibrin begins to appear.
80
are all transcription factors involved in proliferation, responsiveness Somatic Mutations and Interclonal Diversity Myeloma
to IL-6 and BMSC-MM (multiple myeloma) adhesion, promoting evolves in a stepwise process, transforming from MG to smoldering
81
cell-adhesion-mediated drug resistance (CAM-DR), via integrin α /E- myeloma to overt myeloma, where mutations accumulate, conferring
7
cadherin interactions. The prevalences of these three primary IGH either a growth advantage (driver mutations) or are functionally irrel-
82
rearrangements [t(6;14), t(11;14), and t(14;16)] are similar in MG, indi- evant (passenger mutations). So far, more than 300 myeloma patient
cating the need of additional transforming events to precipitate active DNA samples have been sequenced using whole-genome sequencing
myeloma. 83–85 or whole-exome sequencing approaches. 100,108–113 Specifically, 11 genes
Kaushansky_chapter 107_p1733-1772.indd 1736 9/21/15 12:34 PM