Page 672 - Clinical Hematology_ Theory _ Procedures ( PDFDrive )
P. 672
656 PART 8 ■ Fundamentals of Hematological Analysis
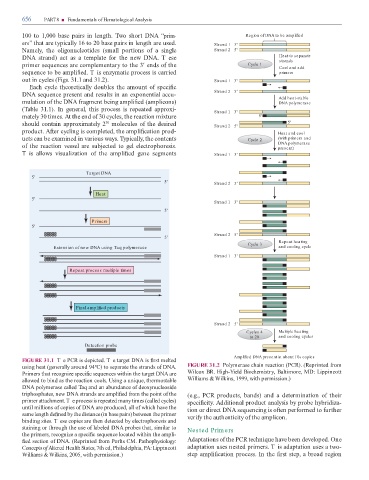
100 to 1,000 base pairs in length. wo short DNA “prim- Region of DNA to be amplified
ers” that are typically 16 to 20 base pairs in length are used. Strand 1 3'
Namely, the oligonucleotides (small portions o a single Strand 2 5'
DNA strand) act as a template or the new DNA. T ese Heat to separate
strands
primer sequences are complementary to the 3′ ends o the Cycle 1 Cool and add
sequence to be ampli ed. T is enzymatic process is carried primers
out in cycles (Figs. 31.1 and 31.2). Strand 1 3'
Each cycle theoretically doubles the amount o speci c
DNA sequence present and results in an exponential accu- Strand 2 5' Add heat-stable
mulation o the DNA ragment being ampli ed (amplicons) DNA polymerase
( able 31.1). In general, this process is repeated approxi- Strand 1 3'
mately 30 times. At the end o 30 cycles, the reaction mixture 5'
should contain approximately 2 molecules o the desired Strand 2 5' 5'
30
product. A er cycling is completed, the ampli cation prod- Heat and cool
ucts can be examined in various ways. ypically, the contents Cycle 2 (with primers and
o the reaction vessel are subjected to gel electrophoresis. DNA polymerase
present)
Tis allows visualization o the ampli ed gene segments Strand 1 3'
Target DNA
5'
5' Strand 2 5'
Heat
5'
Strand 1 3'
5'
Primers
5'
Strand 2 5'
5'
Repeat heating
Cycle 3
Extension of new DNA using Taq polymerase and cooling cycle
Strand 1 3'
Repeat process multiple times
Final amplified products
Strand 2 5'
Cycles 4 Multiple heating
to 20 and cooling cycles
Detection probe
Amplified DNA present in about 106 copies
FIGURE 31.1 Te PCR is depicted. Te target DNA is rst melted
using heat (generally around 94°C) to separate the strands o DNA. FIGURE 31.2 Polymerase chain reaction (PCR). (Reprinted rom
Primers that recognize speci c sequences within the target DNA are Wilcox BR. High-Yield Biochemistry, Baltimore, MD: Lippincott
allowed to bind as the reaction cools. Using a unique, thermostable Williams & Wilkins, 1999, with permission.)
DNA polymerase called aq and an abundance o deoxynucleoside
triphosphates, new DNA strands are ampli ed rom the point o the (e.g., PCR products, bands) and a determination o their
primer attachment. T e process is repeated many times (called cycles) speci city. Additional product analysis by probe hybridiza-
until millions o copies o DNA are produced, all o which have the tion or direct DNA sequencing is o en per ormed to urther
same length de ned by the distance (in base pairs) between the primer veri y the authenticity o the amplicon.
binding sites. T ese copies are then detected by electrophoresis and
staining or through the use o labeled DNA probes that, similar to Nested Prim ers
the primers, recognize a speci c sequence located within the ampli-
ed section o DNA. (Reprinted rom Porlts CM. Pathophysiology: Adaptations o the PCR technique have been developed. One
Concepts o Altered Health States, 7th ed, Philadelphia, PA: Lippincott adaptation uses nested primers. T is adaptation uses a two-
Williams & Wilkins, 2005, with permission.) step ampli cation process. In the rst step, a broad region