Page 675 - Clinical Hematology_ Theory _ Procedures ( PDFDrive )
P. 675
CHAPTER 31 ■ Molecular Diagnostic Techniques and Applications 659
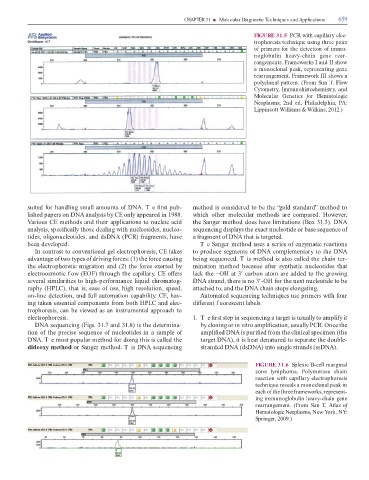
FIGURE 31.5 PCR with capillary elec-
trophoresis technique using three pairs
o primers or the detection o immu-
noglobulin heavy-chain gene rear-
rangements. Frameworks I and II show
a monoclonal peak, representing gene
rearrangement. Framework III shows a
polyclonal pattern. (From Sun . Flow
Cytometry, Immunohistochemistry, and
Molecular Genetics or Hematologic
Neoplasms, 2nd ed, Philadelphia, PA:
Lippincott Williams & Wilkins, 2012.)
suited or handling small amounts o DNA. T e rst pub- method is considered to be the “gold standard” method to
lished papers on DNA analysis by CE only appeared in 1988. which other molecular methods are compared. However,
Various CE methods and their applications to nucleic acid the Sanger method does have limitations (Box 31.3). DNA
analysis, speci cally those dealing with nucleosides, nucleo- sequencing displays the exact nucleotide or base sequence o
tides, oligonucleotides, and dsDNA (PCR) ragments, have a ragment o DNA that is targeted.
been developed. T e Sanger method uses a series o enzymatic reactions
In contrast to conventional gel electrophoresis, CE takes to produce segments o DNA complementary to the DNA
advantage o two types o driving orces: (1) the orce causing being sequenced. T is method is also called the chain ter-
the electrophoretic migration and (2) the orce exerted by mination method because a er synthetic nucleotides that
electroosmotic f ow (EOF) through the capillary. CE o ers lack the −OH at 3′ carbon atom are added to the growing
several similarities to high-per ormance liquid chromatog- DNA strand, there is no 3′-OH or the next nucleotide to be
raphy (HPLC), that is, ease o use, high resolution, speed, attached to, and the DNA chain stops elongating.
on-line detection, and ull automation capability. CE, hav- Automated sequencing techniques use primers with our
ing taken essential components rom both HPLC and elec- di erent f uorescent labels.
trophoresis, can be viewed as an instrumental approach to
electrophoresis. 1. T e rst step in sequencing a target is usually to ampli y it
DNA sequencing (Figs. 31.7 and 31.8) is the determina- by cloning or in vitro ampli cation, usually PCR. Once the
tion o the precise sequence o nucleotides in a sample o ampli ed DNA is puri ed rom the clinical specimen (the
DNA. T e most popular method or doing this is called the target DNA), it is heat denatured to separate the double-
dideoxy method or Sanger method. T is DNA sequencing stranded DNA (dsDNA) into single strands (ssDNA).
FIGURE 31.6 Splenic B-cell marginal
zone lymphoma. Polymerase chain
reaction with capillary electrophoresis
technique reveals a monoclonal peak in
each o the three rameworks, represent-
ing immunoglobulin heavy-chain gene
rearrangement. (From Sun . Atlas o
Hematologic Neoplasms, New York, NY:
Springer, 2009.)