Page 677 - Clinical Hematology_ Theory _ Procedures ( PDFDrive )
P. 677
CHAPTER 31 ■ Molecular Diagnostic Techniques and Applications 661
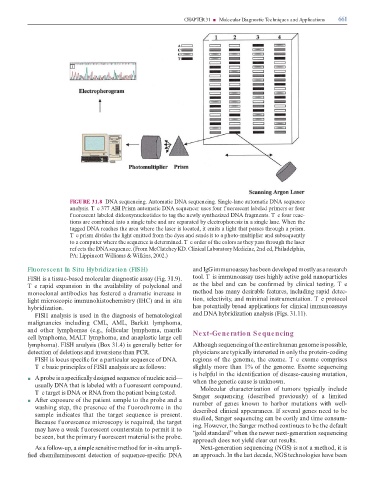
FIGURE 31.8 DNA sequencing. Automatic DNA sequencing. Single-lane automatic DNA sequence
analysis. T e 377 ABI Prism automatic DNA sequencer uses our f uorescent labeled primers or our
f uorescent labeled dideoxynucleotides to tag the newly synthesized DNA ragments. T e our reac-
tions are combined into a single tube and are separated by electrophoresis in a single lane. When the
tagged DNA reaches the area where the laser is located, it emits a light that passes through a prism.
T e prism divides the light emitted rom the dyes and sends it to a photo-multiplier and subsequently
to a computer where the sequence is determined. T e order o the colors as they pass through the laser
ref ects the DNA sequence. (From McClatchey KD. Clinical Laboratory Medicine, 2nd ed, Philadelphia,
PA: Lippincott Williams & Wilkins, 2002.)
Fluorescent In Situ Hybridization (FISH) and IgG immunoassay has been developed mostly as a research
FISH is a tissue-based molecular diagnostic assay (Fig. 31.9). tool. T is immunoassay uses highly active gold nanoparticles
T e rapid expansion in the availability o polyclonal and as the label and can be con rmed by clinical testing. T e
monoclonal antibodies has ostered a dramatic increase in method has many desirable eatures, including rapid detec-
light microscopic immunohistochemistry (IHC) and in situ tion, selectivity, and minimal instrumentation. T e protocol
hybridization. has potentially broad applications or clinical immunoassays
FISH analysis is used in the diagnosis o hematological and DNA hybridization analysis (Figs. 31.11).
malignancies including CML, AML, Burkitt lymphoma,
and other lymphomas (e.g., ollicular lymphoma, mantle Next-Generation Sequencing
cell lymphoma, MAL lymphoma, and anaplastic large cell
lymphoma). FISH analysis (Box 31.4) is generally better or Although sequencing o the entire human genome is possible,
detection o deletions and inversions than PCR. physicians are typically interested in only the protein-coding
FISH is locus speci c or a particular sequence o DNA. regions o the genome, the exome. T e exome comprises
T e basic principles o FISH analysis are as ollows: slightly more than 1% o the genome. Exome sequencing
is help ul in the identi cation o disease-causing mutation,
■ A probe is a speci cally designed sequence o nucleic acid— when the genetic cause is unknown.
usually DNA that is labeled with a f uorescent compound. Molecular characterization o tumors typically include
Te target is DNA or RNA rom the patient being tested. Sanger sequencing (described previously) o a limited
■ A er exposure o the patient sample to the probe and a number o genes known to harbor mutations with well-
washing step, the presence o the f uorochrome in the described clinical appearances. I several genes need to be
sample indicates that the target sequence is present. studied, Sanger sequencing can be costly and time consum-
Because f uorescence microscopy is required, the target ing. However, the Sanger method continues to be the de ault
may have a weak f uorescent counterstain to permit it to “gold standard” when the newer next-generation sequencing
be seen, but the primary f uorescent material is the probe.
approach does not yield clear cut results.
As a ollow-up, a simple sensitive method or in-situ ampli- Next-generation sequencing (NGS) is not a method, it is
ed chemiluminescent detection o sequence-speci c DNA an approach. In the last decade, NGS technologies have been