Page 1364 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 1364
1210 Part VII Hematologic Malignancies
EBV
infection
(40%) CD40 BCMA TACI
RANK
CD40 LMP1
CD30
TRAF3
RIP TRAF NIK gains mutations
TNFAIP3 (20%) NIK TRAF3
mutations TNFAIP3 CYLD (5%)
(40%)
CYLD
Classical NEMO mutations
NF-κB IKKα IKKβ (5%) IKKα IKKα Alternative
pathway NF-κB
pathway
NFKBIA and
NFKBIE mutations IκBα/
(10–20%) IκBε p100 RELB
p50 p65 Proteasomal
degradation
REL
amplification Cytoplasm
(40%) BCL3 gains or
translocations
(rare) p52 RELB Nucleus
p50 p65 BCL3
p50 p50
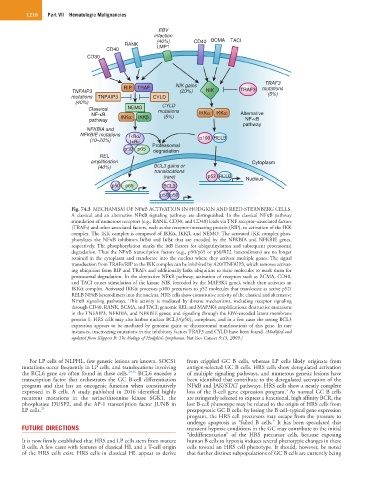
Fig. 74.3 MECHANISM OF NFκB ACTIVATION IN HODGKIN AND REED-STERNBERG CELLS.
A classical and an alternative NFκB signaling pathway are distinguished. In the classical NFκB pathway
stimulation of numerous receptors (e.g., RANK, CD30, and CD40) leads via TNF receptor–associated factors
(TRAFs) and other associated factors, such as the receptor-interacting protein (RIP), to activation of the IKK
complex. The IKK complex is composed of IKKα, IKKβ, and NEMO. The activated IKK complex phos-
phorylates the NFκB inhibitors IκBα and IκBε that are encoded by the NFKBIA and NFKBIE genes,
respectively. The phosphorylation marks the IκB factors for ubiquitinylation and subsequent proteasomal
degradation. Thus the NFκB transcription factors (e.g., p50/p65 or p50/REL heterodimers) are no longer
retained in the cytoplasm and translocate into the nucleus where they activate multiple genes. The signal
transduction from TRAFs/RIP to the IKK complex can be inhibited by A20/TNFAIP3, which removes activat-
ing ubiquitins from RIP and TRAFs and additionally links ubiquitins to these molecules to mark them for
proteasomal degradation. In the alternative NFκB pathway, activation of receptors such as BCMA, CD40,
and TACI causes stimulation of the kinase NIK (encoded by the MAP3K4 gene), which then activates an
IKKα complex. Activated IKKα processes p100 precursors to p52 molecules that translocate as active p52/
RELB NFκB heterodimers into the nucleus. HRS cells show constitutive activity of the classical and alternative
NFκB signaling pathways. This activity is mediated by diverse mechanisms, including receptor signaling
through CD40, RANK, BCMA, and TACI; genomic REL and MAP3K4 amplifications; destructive mutations
in the TNFAIP3, NFKBIA, and NFKBIE genes; and signaling through the EBV-encoded latent membrane
protein 1. HRS cells may also harbor nuclear BCL3/(p50) 2 complexes, and in a few cases the strong BCL3
expression appears to be mediated by genomic gains or chromosomal translocations of this gene. In rare
instances, inactivating mutations in the inhibitory factors TRAF3 and CYLD have been found. (Modified and
updated from Küppers R: The biology of Hodgkin’s lymphoma. Nat Rev Cancer 9:15, 2009.)
For LP cells of NLPHL, few genetic lesions are known. SOCS1 from crippled GC B cells, whereas LP cells likely originate from
mutations occur frequently in LP cells, and translocations involving antigen-selected GC B cells. HRS cells show deregulated activation
the BCL6 gene are often found in these cells. 29,30 BCL6 encodes a of multiple signaling pathways, and numerous genetic lesions have
transcription factor that orchestrates the GC B-cell differentiation been identified that contribute to the deregulated activation of the
program and that has an oncogenic function when constitutively NFκB and JAK/STAT pathways. HRS cells show a nearly complete
3
expressed in B cells. A study published in 2016 identified highly loss of the B-cell gene expression program. As normal GC B cells
recurrent mutations in the serine/threonine kinase SGK1, the are stringently selected to express a functional, high affinity BCR, the
phosphatase DUSP2, and the AP-1 transcription factor JUNB in lost B-cell phenotype may be related to the origin of HRS cells from
LP cells. 29 preapoptotic GC B cells: by losing the B cell–typical gene expression
program, the HRS cell precursors may escape from the pressure to
undergo apoptosis as “failed B cells.” It has been speculated that
FUTURE DIRECTIONS transient hypoxic conditions in the GC may contribute to the initial
“dedifferentiation” of the HRS precursor cells, because exposing
It is now firmly established that HRS and LP cells stem from mature human B cells to hypoxia induces several phenotypic changes in these
B cells. A few cases with features of classical HL and a T-cell origin cells toward an HRS cell phenotype. It should, however, be noted
of the HRS cells exist. HRS cells in classical HL appear to derive that further distinct subpopulations of GC B cells are currently being