Page 1393 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 1393
Chapter 76 Origin of Non-Hodgkin Lymphoma 1239
Although BCL2 is thought to have a critical role in development evolution or transformation of a putative progenitor FL stem cell.
of FL, nearly 30% of grade 3A and a majority of grade 3B FLs are Acquired rearrangement of the MYC oncogene can also occur and
t(14;18) negative. Alternative rearrangements involving the κ or λ has been associated with an aggressive plasmablastic phenotype.
light chains and BCL2 may occur, but many t(14;18) FLs do not Stromal factors with FL tumors also likely play a key role in FL
express BCL2. These entities express a post-GC CD10-IRF4-pheno- propagation and have been studied as having prognostic consequence
type with 3q27/BCL6 rearrangements and are associated with lower in FL as well.
chemotherapy response rates as well as worse overall survival com-
pared with t(14;18)-positive FLs. It is not clear how this subset of FL
evolves from normal B cells, but this immunophenotype suggests a Tumor Microenvironment and Survival in
late- or post-GC origin. GEP and analysis of miRNA profiles of FL Follicular Lymphoma
confirm this association. Leich et al analyzed gene expression profiles
of 184 grade I-3A FLs, of which 17 were t(14;18) negative (six of Just as the lymphoid microenvironment is integral to the formation
these rearrangement-negative FLs still overexpressed BCL2). Analysis and function of GCs and immune factors predict survival in DLBCL,
of this data showed enrichment of signatures associated with activated FL development appears to depend on stromal factors and the host
B cells, including NFκB signaling, and those associated with cell immune response. In the largest study examining this relationship
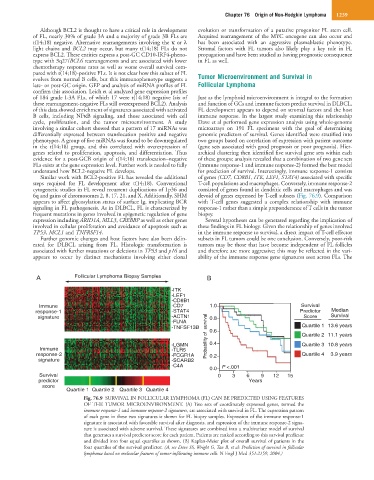
cycle, proliferation, and the tumor microenvironment. A study Dave et al performed gene expression analysis using whole-genome
involving a similar cohort showed that a pattern of 17 miRNAs was microarrays on 191 FL specimens with the goal of determining
differentially expressed between translocation positive and negative genomic predictors of survival. Genes identified were stratified into
phenotypes. A group of five miRNAs was found to be downregulated two groups based on correlation of expression with patient outcome
in the t(14;18) group, and this correlated with overexpression of (gene sets associated with good prognosis or poor prognosis). Hier-
genes related to proliferation, apoptosis, and differentiation. Thus archical clustering then identified five survival gene sets within each
evidence for a post-GCB origin of t(14;18) translocation–negative of these groups; analysis revealed that a combination of two gene sets
FLs exists at the gene expression level. Further work is needed to fully (immune response-1 and immune response-2) formed the best model
understand how BCL2-negative FL develops. for prediction of survival. Interestingly, immune response-1 consists
Similar work with BCL2-positive FL has revealed the additional of genes (CD7, CD8B1, ITK, LEF1, STAT4) associated with specific
steps required for FL development after t(14;18). Conventional T-cell populations and macrophages. Conversely, immune response-2
cytogenetic studies in FL reveal recurrent duplications of 1p36 and consisted of genes found in dendritic cells and macrophages and was
6q and gains of chromosomes 2, 8, 17, 21, and X. Additionally, SHM devoid of genes expressed by T-cell subsets (Fig. 76.9). Comparison
appears to affect glycosylation status of surface Ig, implicating BCR with T-cell genes suggested a complex relationship with immune
signaling in FL pathogenesis. As in DLBCL, FL is characterized by response-1 rather than a simple preponderance of T cells in the tumor
frequent mutations in genes involved in epigenetic regulation of gene biopsy.
expression including ARID1A, MLL3, CREBBP as well as other genes Several hypotheses can be generated regarding the implication of
involved in cellular proliferation and avoidance of apoptosis such as these findings in FL biology. Given the relationship of genes involved
TP53, MCL1 and TNFRSF14. in the immune response to survival, a direct impact of T-cell effector
Further genomic changes and host factors have also been delin- subsets in FL tumors could be one conclusion. Conversely, poor-risk
eated for DLBCL arising from FL. Histologic transformation is tumors may be those that have become independent of FL follicles
associated with further mutations or deletions in TP53 and p16 and and therefore are more aggressive; this may be reflected in the vari-
appears to occur by distinct mechanisms involving either clonal ability of the immune response gene signatures seen across FLs. The
A Follicular Lymphoma Biopsy Samples B
-ITK
-LEF1
-CD8B1
Immune -CD7 1.0 Survival
response-1 -STAT4 Predictor Median
signature -ACTN1 0.8 Score Survival
-FLNA
-TNFSF13B 0.6 Quartile 1 13.6 years
Quartile 2 11.1 years
-LGMN Probability of survival 0.4 Quartile 3 10.8 years
Immune -TLR5
response-2 -FCGR1A 0.2 Quartile 4 3.9 years
signature -SCARB2
-C4A P <.001
0.0
Survival 0 3 6 9 12 15
predictor Years
score
Quartile 1 Quartile 2 Quartile 3 Quartile 4
Fig. 76.9 SURVIVAL IN FOLLICULAR LYMPHOMA (FL) CAN BE PREDICTED USING FEATURES
OF THE TUMOR MICROENVIRONMENT. (A) Two sets of coordinately expressed genes, termed the
immune response-1 and immune response-2 signatures, are associated with survival in FL. The expression pattern
of each gene in these two signatures is shown for FL biopsy samples. Expression of the immune response-1
signature is associated with favorable survival after diagnosis, and expression of the immune response-2 signa-
ture is associated with adverse survival. These signatures are combined into a multivariate model of survival
that generates a survival predictor score for each patient. Patients are ranked according to this survival predictor
and divided into four equal quartiles as shown. (B) Kaplan-Meier plot of overall survival of patients in the
four quartiles of the survival predictor. (A, see Dave SS, Wright G, Tan B, et al: Prediction of survival in follicular
lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med 351:2159, 2004.)