Page 895 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 895
778 Part VII Hematologic Malignancies
abnormalities in nondividing, interphase nuclei (Fig. 56.5). Six
aspects of interphase FISH are particularly useful: (1) Interphase
cytogenetics allows screening of a large number of cells. This permits
investigation of hematologic malignancies with a low mitotic yield,
such as chronic lymphocytic leukemia (CLL) or multiple myeloma
(MM). (2) Interphase FISH permits detection of chromosomal rear-
rangements in peripheral blood samples, thus obviating the need for
marrow aspiration. For instance, in CML, which rarely yields a large
number of dividing cells in peripheral blood, conventional cytogenet-
A ics usually is uninformative. However, detection of BCR-ABL1, a
molecular equivalent of the Philadelphia chromosome (Ph), in
peripheral blood using interphase FISH provides reliable, fast, quan-
titative results (see the section on Chronic Myelogenous Leukemia
later in this chapter). (3) Interphase FISH offers a quantitative assay
for monitoring disease progression or detection of minimal residual
disease after ablative chemotherapy or hematopoietic stem cell
transplantation. (4) Use of specific probe sets allows detection of
specific disease-associated abnormalities such as t(8;21), which
B denotes the M2 subtype of acute myeloid leukemia (AML), or
t(15;17), which is associated with acute promyelocytic leukemia
(APL), within 4 hours, allowing for timely and appropriate therapy.
(5) Abnormalities can be detected accurately in archival specimens
stored for up to 15 years. (6) Simultaneous use of interphase FISH
and immunophenotyping is a powerful tool for investigation of
lineage involvement in diseases such as myelodysplasia and to deter-
mine which cell population carries the specific chromosome abnor-
C mality. FISH nomenclature is described in the International System
for Human Cytogenetic Nomenclature.
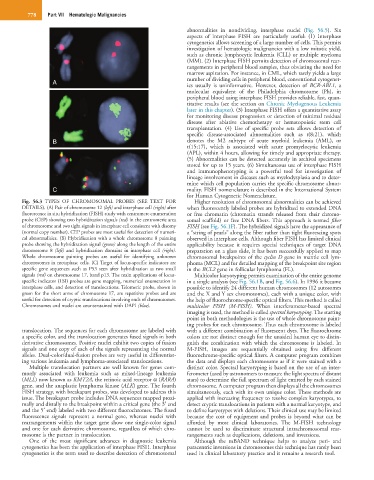
Fig. 56.3 TYPES OF CHROMOSOMAL PROBES (SEE TEXT FOR Higher resolution of chromosomal abnormalities can be achieved
DETAILS). (A) Pair of chromosome 12 (left) and interphase cell (right) after when fluorescently labeled probes are hybridized to extended DNA
fluorescence in situ hybridization (FISH) study with centromere enumeration or free chromatin (chromatin strands released from their chromo-
probe (CEP) showing two hybridization signals (red) in the centromeric area somal scaffold) or free DNA fibers. This approach is termed fiber
of chromosome and two tight signals in interphase cell consistent with disomy FISH (see Fig. 56.1F). The hybridized signals have the appearance of
(normal copy number). CEP probes are most useful for detection of numeri- a “string of pearls” along the fiber rather than tight fluorescing spots
cal abnormalities. (B) Hybridization with a whole chromosome 8 painting observed in interphase cells. Although fiber FISH has limited clinical
probe showing the hybridization signal (green) along the length of the entire applicability because it requires special techniques of target DNA
chromosome 8 (left) and hybridization domains in interphase cell (right). preparation on a glass slide, it has been successfully applied to map
Whole chromosome painting probes are useful for identifying unknown chromosomal breakpoints of the cyclin D gene in mantle cell lym-
chromosomes in metaphase cells. (C) Target of locus-specific indicators are phoma (MCL) and for detailed mapping of the breakpoint site region
specific gene sequences such as P53 seen after hybridization as two small in the BCL2 gene in follicular lymphoma (FL).
signals (red) on chromosome 17, band p13. The main applications of locus- Multicolor karyotyping permits examination of the entire genome
specific indicator (LSI) probes are gene mapping, numerical enumeration in in a single analysis (see Fig. 56.1B, and Fig. 56.6). In 1996 it became
interphase cells, and detection of translocations. Telomeric probe, shown in possible to identify 24 different human chromosomes (12 autosomes
green for the short arms of chromosome 17, are repetitive probes and are and the X and Y sex chromosomes), each with a unique color, with
useful for detection of cryptic translocations involving ends of chromosomes. the help of fluorochrome-specific optical filters. This method is called
Chromosomes and nuclei are counterstained with DAPI (blue). multicolor FISH (M-FISH). When interferometer-based spectral
imaging is used, the method is called spectral karyotyping. The starting
point in both methodologies is the use of whole chromosome paint-
ing probes for each chromosome. Thus each chromosome is labeled
translocation. The sequences for each chromosome are labeled with with a different combination of fluorescent dyes. The fluorochrome
a specific color, and the translocation generates fused signals in both colors are not distinct enough for the unaided human eye to distin-
derivative chromosomes. Positive nuclei exhibit two copies of fusion guish the combination with which the chromosome is labeled. In
signals and one copy of each of the signals representing the normal M-FISH, images are sequentially obtained using five different
alleles. Dual-color/dual-fusion probes are very useful in differentiat- fluorochrome-specific optical filters. A computer program combines
ing various leukemia and lymphoma-associated translocations. the data and displays each chromosome as if it were stained with a
Multiple translocation partners are well known for genes com- distinct color. Spectral karyotyping is based on the use of an inter-
monly associated with leukemia such as mixed-lineage leukemia ferometer (used by astronomers to measure the light spectra of distant
(MLL) now known as KMT2A, the retinoic acid receptor α (RARA) stars) to determine the full spectrum of light emitted by each stained
gene, and the anaplastic lymphoma kinase (ALK) gene. The fourth chromosome. A computer program then displays all the chromosomes
FISH strategy, with breakapart probes, was developed to address this simultaneously, each with its own unique color. These methods are
issue. The breakapart probe includes DNA sequences mapped proxi- applied with increasing frequency to resolve complex karyotypes, to
mally and distally to the breakpoint within a critical gene (the 3′ end detect cryptic translocations in patients with a normal karyotype, and
and the 5′ end) labeled with two different fluorochromes. The fused to define karyotypes with deletions. Their clinical use may be limited
fluorescence signals represent a normal gene, whereas nuclei with because the cost of equipment and probes is beyond what can be
rearrangements within the target gene show one single-color signal afforded by most clinical laboratories. The M-FISH technology
and one for each derivative chromosome, regardless of which chro- cannot be used to discriminate structural intrachromosomal rear-
mosome is the partner in translocation. rangements such as duplications, deletions, and inversions.
One of the most significant advances in diagnostic leukemia Although the mBAND technique helps to analyze peri- and
cytogenetics has been the application of interphase FISH. Interphase paracentric inversions in chromosomes this technique has rarely been
cytogenetics is the term used to describe detection of chromosomal used in clinical laboratory practice and it remains a research tool.