Page 894 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 894
Chapter 56 Conventional and Molecular Cytogenomic Basis of Hematologic Malignancies 777
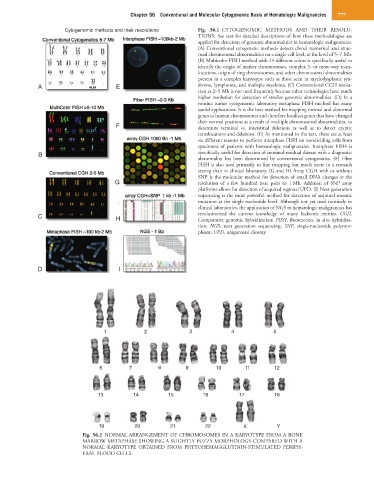
Cytogenomic methods and their resolutions Fig. 56.1 CYTOGENOMIC METHODS AND THEIR RESOLU-
TIONS. See text for detailed descriptions of how these methodologies are
applied for detection of genomic abnormalities in hematologic malignancies.
(A) Conventional cytogenetic methods detects clonal numerical and struc-
tural chromosomal abnormalities on a single-cell level, at the level of 5–7 Mb.
(B) Multicolor FISH method with 24 different colors is specifically useful to
identify the origin of marker chromosomes, complex 3- or more-way trans-
locations, origin of ring chromosomes, and other chromosomal abnormalities
present in a complex karyotype such as those seen in myelodysplastic syn-
A E drome, lymphoma, and multiple myeloma. (C) Conventional CGH resolu-
tion at 2–5 Mb is not used frequently because other technologies have much
higher resolution for detection of smaller genomic abnormalities. (D) In a
routine tumor cytogenomic laboratory metaphase FISH method has many
useful applications. It is the best method for mapping normal and abnormal
genes to human chromosomes and therefore localizes genes that have changed
their normal positions as a result of multiple chromosomal abnormalities, to
F determine terminal vs. interstitial deletions as well as to detect cryptic
translocations and deletions. (E) As mentioned in the text, there are at least
six different reasons to perform interphase FISH on nondividing cells from
specimens of patients with hematologic malignancies. Interphase FISH is
B specifically useful for detection of minimal-residual disease with a diagnostic
abnormality has been determined by conventional cytogenetics. (F) Fiber
FISH is also used primarily in fine mapping but much more in a research
setting than in clinical laboratory. (G and H) Array CGH with or without
SNP is the molecular method for detection of small DNA changes at the
G resolution of a few hundred base pairs to 1 Mb. Addition of SNP array
platforms allows for detection of acquired regional UPD. (I) Next generation
sequencing is the most powerful method for detection of acquired somatic
mutation at the single nucleotide level. Although not yet used routinely in
clinical laboratories, the application of NGS to hematologic malignancies has
C H revolutionized the current knowledge of many leukemic entities. CGH,
Comparative genomic hybridization; FISH, fluorescence in situ hybridiza-
tion; NGS, next generation sequencing; SNP, single-nucleotide polymor-
phism; UPD, uniparental disomy.
D I
1 2 3 4 5
6 7 8 9 10 11 12
13 14 15 16 17 18
19 20 21 22 X Y
Fig. 56.2 NORMAL ARRANGEMENT OF CHROMOSOMES IN A KARYOTYPE FROM A BONE
MARROW METAPHASE SHOWING A SLIGHTLY FUZZY MORPHOLOGY COMPARED WITH A
NORMAL KARYOTYPE OBTAINED FROM PHYTOHEMAGGLUTININ-STIMULATED PERIPH-
ERAL BLOOD CELLS.