Page 898 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 898
Chapter 56 Conventional and Molecular Cytogenomic Basis of Hematologic Malignancies 781
1 2 3 4 5
6 7 8 9 10 11 12
13 14 15 16 17 18
19 20 21 22 X Y
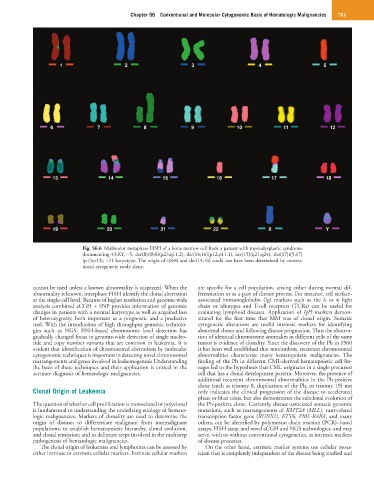
Fig. 56.6 Multicolor metaphase FISH of a bone marrow cell from a patient with myelodysplastic syndrome
documenting 43,XY, −5, der(8)t(8;8)(p23;q11.2), der(14;16)(p12;p11.1), inv(15)(q21;q24), der(17)t(5;17)
(p13;p13), −21 karyotype. The origin of t(8;8) and der(14;16) could not have been determined by conven-
tional cytogenetic study alone.
cannot be used unless a known abnormality is suspected. When the are specific for a cell population, arising either during normal dif-
abnormality is known, interphase FISH identify the clonal aberration ferentiation or as a part of disease process. For instance, cell surface-
at the single-cell level. Because of higher resolution and genome-wide associated immunoglobulin (Ig) markers such as the λ or κ light
analysis combined aCGH + SNP provides information of genomic chain or idiotypes and T-cell receptors (TCRs) can be useful for
changes in patients with a normal karyotype as well as acquired loss evaluating lymphoid diseases. Application of IgH markers demon-
of heterozygosity, both important as a prognostic and a predictive strated for the first time that MM was of clonal origin. Somatic
tool. With the introduction of high throughput genomic technolo- cytogenetic alterations are useful intrinsic markers for identifying
gies such as NGS, FISH-based chromosome level detection has abnormal clones and following disease progression. Thus the observa-
gradually changed focus to genome-wide detection of single nucleo- tion of identical chromosome anomalies in different cells of the same
tide and copy number variants that are common in leukemia. It is tumor is evidence of clonality. Since the discovery of the Ph in 1960
evident that identification of chromosomal aberrations by molecular it has been well established that nonrandom, recurrent chromosomal
cytogenomic techniques is important in detecting novel chromosomal abnormalities characterize many hematopoietic malignancies. The
rearrangements and genes involved in leukemogenesis Understanding finding of the Ph in different CML-derived hematopoietic cell lin-
the basis of these techniques and their application is critical in the eages led to the hypothesis that CML originates in a single precursor
accurate diagnosis of hematologic malignancies. cell that has a clonal development pattern. Moreover, the presence of
additional recurrent chromosomal abnormalities in the Ph-positive
clone (such as trisomy 8, duplication of the Ph, or trisomy 19) not
Clonal Origin of Leukemia only indicates the clinical progression of the disease to accelerated
phase or blast crisis, but also demonstrates the subclonal evolution of
The question of whether cell proliferation is monoclonal or polyclonal the Ph-positive clone. Currently, disease-associated somatic genomic
is fundamental to understanding the underlying etiology of hemato- mutations, such as rearrangements of KMT2A (MLL), runt-related
logic malignancies. Markers of clonality are used to determine the transcription factor gene (RUNX1), ETV6, PML-RARA, and many
origin of disease; to differentiate malignant from nonmalignant others, can be identified by polymerase chain reaction (PCR)–based
populations; to establish hematopoietic hierarchy, clonal evolution, assays, FISH assay, and novel aCGH and NGS technologies, and may
and clonal remission; and to delineate steps involved in the multistep serve, with or without conventional cytogenetics, as intrinsic markers
pathogenesis of hematologic malignancies. of disease processes.
The clonal origin of leukemias and lymphomas can be assessed by On the other hand, extrinsic marker systems use cellular mosa-
either intrinsic or extrinsic cellular markers. Intrinsic cellular markers icism that is completely independent of the disease being studied and