Page 893 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 893
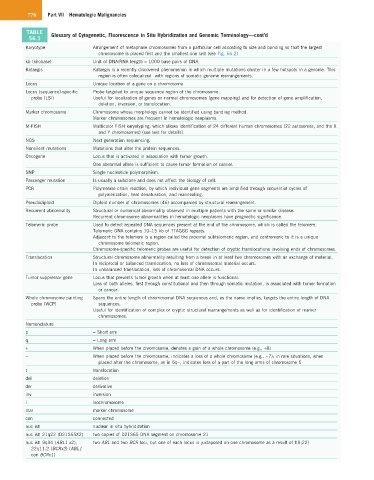
776 Part VII Hematologic Malignancies
TABLE Glossary of Cytogenetic, Fluorescence in Situ Hybridization and Genomic Terminology—cont’d
56.1
Karyotype Arrangement of metaphase chromosomes from a particular cell according to size and banding so that the largest
chromosome is placed first and the smallest one last (see Fig. 56.2).
kb (kilobase) Unit of DNA/RNA length = 1000 base pairs of DNA.
Kataegis Kataegis is a recently discovered phenomenon in which multiple mutations cluster in a few hotspots in a genome. This
region is often colocalized with regions of somatic genome rearrangements.
Locus Unique location of a gene on a chromosome.
Locus (sequence)-specific Probe targeted to unique sequence region of the chromosome.
probe (LSI) Useful for localization of genes on normal chromosomes (gene mapping) and for detection of gene amplification,
deletion, inversion, or translocation.
Marker chromosome Chromosome whose morphology cannot be identified using banding method.
Marker chromosomes are frequent in hematologic neoplasms.
M-FISH Multicolor FISH karyotyping, which allows identification of 24 different human chromosomes (22 autosomes, and the X
and Y chromosomes) (see text for details).
NGS Next generation sequencing.
Nonsilent mutations Mutations that alter the protein sequences.
Oncogene Locus that is activated in association with tumor growth.
One abnormal allele is sufficient to cause tumor formation or cancer.
SNP Single nucleotide polymorphism.
Passenger mutation Is usually a subclone and does not affect the biology of cell.
PCR Polymerase chain reaction, by which individual gene segments are amplified through sequential cycles of
polymerization, heat denaturation, and reannealing.
Pseudodiploid Diploid number of chromosomes (46) accompanied by structural rearrangement.
Recurrent abnormality Structural or numerical abnormality observed in multiple patients with the same or similar disease.
Recurrent chromosome abnormalities in hematologic neoplasms have prognostic significance.
Telomeric probe Used to detect repeated DNA sequences present at the end of the chromosome, which is called the telomere.
Telomeric DNA contains 10–15 kb of TTAGGG repeats.
Adjacent to the telomere is a region called the proximal subtelomeric region, and centromeric to it is a unique
chromosome telomeric region.
Chromosome-specific telomeric probes are useful for detection of cryptic translocations involving ends of chromosomes.
Translocation Structural chromosome abnormality resulting from a break in at least two chromosomes with an exchange of material.
In reciprocal or balanced translocation, no loss of chromosomal material occurs.
In unbalanced translocation, loss of chromosomal DNA occurs.
Tumor suppressor gene Locus that prevents tumor growth when at least one allele is functional.
Loss of both alleles, first through constitutional and then through somatic mutation, is associated with tumor formation
or cancer.
Whole chromosome painting Spans the entire length of chromosomal DNA sequences and, as the name implies, targets the entire length of DNA
probe (WCP) sequences.
Useful for identification of complex or cryptic structural rearrangements as well as for identification of marker
chromosomes.
Nomenclature
p = Short arm
q = Long arm
+ When placed before the chromosome, denotes a gain of a whole chromosome (e.g., +8)
− When placed before the chromosome, indicates a loss of a whole chromosome (e.g., −7); in rare situations, when
placed after the chromosome, as in 5q−, indicates loss of a part of the long arms of chromosome 5
t translocation
del deletion
der derivative
inv inversion
i isochromosome
mar marker chromosome
con connected
nuc ish nuclear in situ hybridization
nuc ish 21q22 (D21S65X2) two copies of D21S65 DNA segment on chromosome 21
nuc ish 9q34 (ABL1 x2), two ABL and two BCR loci, but one of each locus is juxtaposed on one chromosome as a result of t(9;22)
22q11.2 (BCRx2) (ABL1
con BCRx1)