Page 921 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 921
804 Part VII Hematologic Malignancies
X = PML (15q22)
MLL (11q23) RARA (17q21)
PLZF/ZBTB16 (11q23)
NPM (5q35)
NuMA (11q13)
STAT5B (17q11)
PRKAR1A (17q24)
FIP1L1 (4q12)
BCOR (Xq11)
Fusion Product
Responsive to ATRA
PML, NPM, NuMA, Stat5b, 3p25
Two syndromes
Nonresponsive to ATRA
PLZF
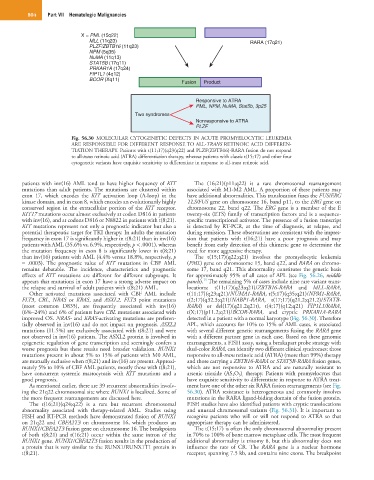
Fig. 56.30 MOLECULAR CYTOGENETIC DEFECTS IN ACUTE PROMYELOCYTIC LEUKEMIA
ARE RESPONSIBLE FOR DIFFERENT RESPONSE TO ALL-TRANS RETINOIC ACID DIFFEREN-
TIATION THERAPY. Patients with t(11;17)(q23(q22) and PLZF(ZBTB16)-RARA fusion do not respond
to all-trans retinoic acid (ATRA) differentiation therapy, whereas patients with classic t(15;17) and other four
cytogenetic variants have exquisite sensitivity to differentiate in response to all-trans retinoic acid.
patients with inv(16) AML tend to have higher frequency of KIT The (16;21)(p11;q22) is a rare chromosomal rearrangement
mutations than adult patients. The mutations are clustered within associated with M1-M2 AML. A proportion of these patients may
exon 17, which encodes the KIT activation loop (A-loop) in the have additional abnormalities. This translocation fuses the FUS/ERG
kinase domain, and in exon 8, which encodes an evolutionarily highly TLS/FUS gene on chromosome 16, band p11, to the ERG gene on
conserved region in the extracellular portion of the KIT receptor. chromosome 22, band q22. The ERG gene is a member of the E
KIT17 mutations occur almost exclusively at codon D816 in patients twenty-six (ETS) family of transcription factors and is a sequence-
with inv(16), and at codons D816 or N8822 in patients with t(8;21). specific transcriptional activator. The presence of a fusion transcript
KIT mutations represent not only a prognostic indicator but also a is detected by RT-PCR, at the time of diagnosis, at relapse, and
potential therapeutic target for TKI therapy. In adults the mutation during remission. These observations are consistent with the impres-
frequency in exon 17 is significantly higher in t(8;21) than in inv(16) sion that patients with t(16;21) have a poor prognosis and may
patients with AML (35.6% vs. 6.9%, respectively, p < .0001), whereas benefit from early detection of this chimeric gene to determine the
the mutation frequency in exon 8 is significantly lower in t(8;21) need for more aggressive therapy.
than inv(16) patients with AML (4.4% versus 18.8%, respectively, p The t(15;17)(q22;q21) involves the promyelocytic leukemia
= .0003). The prognostic value of KIT mutations in CBF AML (PML) gene on chromosome 15, band q22, and RARA on chromo-
remains debatable. The incidence, characteristics and prognostic some 17, band q21. This abnormality constitutes the genetic basis
effects of KIT mutations are different for different subgroups. It for approximately 95% of all cases of APL (see Fig. 56.26, middle
15
appears that mutations in exon 17 have a strong adverse impact on panels). The remaining 5% of cases include nine rare variant trans-
the relapse and survival of adult patients with t(8;21) AML. locations: t(11;17)(q23;q21)/ZBTB16-RARA and MLL-RARA,
Other activated mutations associated with CBF AML include t(11;17)(q23;q21)/NUMA1-RARA, t(5;17)(q35;q21)/NPM1-RARA,
FLT3, CBL, NRAS or KRAS, and ASXL2. FLT3 point mutations t(2;17)(q32.3;q21)/NABP1-RARA, t(17;17)(q21.2;q21.2)/STATB-
(most common D835), are frequently associated with inv(16) RARA) or del(17)(q21.2q21t), t(4;17)(q12;q21) FIP1L1/RARA,
(6%–24%) and 6% of patients have CBL mutations associated with t(X;17)(p11.2;q21)/BCOR-RARA, and cryptic PRKAR1A-RARA
improved OS. NRAS- and KRAS-activating mutations are preferen- detected in a patient with a normal karyotype (Fig. 56.30). Therefore
tially observed in inv(16) and do not impact on prognosis. ASXL2 APL, which accounts for 10% to 15% of AML cases, is associated
mutations (11.5%) are exclusively associated with t(8;21) and were with several different genetic rearrangements fusing the RARA gene
not observed in inv(16) patients. The ASXL2 protein is involved in with a different partner gene in each case. Based on these genomic
epigenetic regulation of gene transcription and seemingly confers a rearrangements, a FISH assay, using a breakapart probe strategy with
worse prognosis but these results need broader validation. RUNX1 dual-color RARA, can identify two different clinical syndromes: those
mutations present in about 5% to 15% of patients with M0 AML, responsive to all-trans retinoic acid (ATRA) (more than 99%) therapy
are mutually exclusive when t(8;21) and inv(16) are present. Approxi- and those carrying a ZBTB16-RARA or STAT5B-RARA fusion genes,
mately 5% to 10% of CBF AML patients, mostly those with t(8;21), which are not responsive to ATRA and are naturally resistant to
have concurrent systemic mastocytosis with KIT mutations and a arsenic trioxide (AS 2 O 3 ) , therapy. Patients with promyelocytes that
good prognosis. have exquisite sensitivity to differentiate in response to ATRA treat-
As mentioned earlier, there are 39 recurrent abnormalities involv- ment have one of the other six RARA fusion rearrangements (see Fig.
ing the 21q22 chromosomal site where RUNX1 is localized. Some of 56.30). ATRA resistance is heterogeneous and commonly involves
the more frequent rearrangements are discussed here. mutations in the RARA ligand-biding domain of the fusion protein.
The t(16;21)(q24;q22) is a rare but recurrent chromosomal FISH studies have also identified patients with cryptic translocations
abnormality associated with therapy-related AML. Studies using and unusual chromosomal variants (Fig. 56.31). It is important to
FISH and RT-PCR methods have demonstrated fusion of RUNX1 recognize patients who will or will not respond to ATRA so that
on 21q22 and CBFA2T3 on chromosome 16, which produces an appropriate therapy can be administered.
RUNX1/CBFA2T3 fusion gene on chromosome 16. The breakpoints The t(15;17) is often the only chromosomal abnormality present
of both t(8;21) and t(16;21) occur within the same intron of the in 70% to 100% of bone marrow metaphase cells. The most frequent
RUNX1 gene. RUNX1/CBFA2T3 fusion results in the production of additional abnormality is trisomy 8, but this abnormality does not
a protein that is very similar to the RUNX1/RUNX1T1 protein in influence the rate of CR. The RARA gene is a nuclear hormone
t(8;21). receptor, spanning 7.5 kb, and contains nine exons. The breakpoint