Page 918 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 918
Chapter 56 Conventional and Molecular Cytogenomic Basis of Hematologic Malignancies 801
t(15;17)/PML-RARA 11%
Other adverse 14%
t(8;21)RUNX1-RUNX1-T1
RUNX1 mut 4% 8%
MLL-PTD 5%
inv(16)/CBFB-MYH11
inv(3)t(3;3)/EVI-1 3% 6%
ADVERSE
26% FAVORABLE
45%
FLT3-ITD/NPM1 wt 12% NPM1 Mut/FLT3-ITD
neg/WT1
ADVERSE wt 18%
17%
Intermediate 17% CEBPAmut (biallelic)/FLT3-
ITD neg 3%
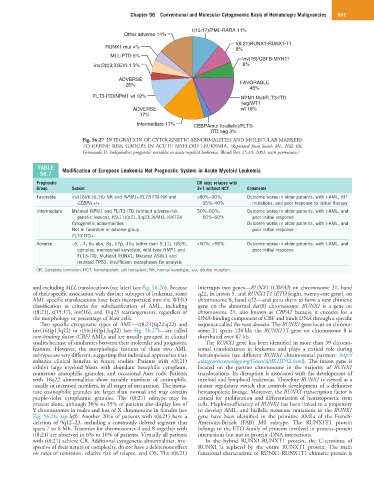
Fig. 56.27 INTEGRATION OF CYTOGENETIC ABNORMALITIES AND MOLECULAR MARKERS
TO REFINE RISK GROUPS IN ACUTE MYELOID LEUKEMIA. (Reprinted from Smith ML, Hills RK,
Grimwade D: Independent prognostic variables in acute myeloid leukemia. Blood Rev 25:39, 2001; with permission.)
TABLE Modification of European Leukemia Net Prognostic System in Acute Myeloid Leukemia
56.7
Prognostic CR rate; relapse with
Group Subset 3+7 without HCT Comments
Favorable inv(16)/t(16;16) NK and NPM1+/FLT3 ITD-NK and >80%–90%; Outcome worse in older patients, with t-AML, KIT
CEBPA +/+ 35%–40% mutations, and poor response to initial therapy
Intermediate Mutated NPM1 and FLIT3-ITD (without adverse-risk 50%–80%; Outcome worse in older patients, with t-AML, and
genetic lession), t(9;11)(p21.3;q23.3)/MLL-KMT2A 50%–60% poor initial response
Cytogenetic abnormalities Outcome worse in older patients, with t-AML, and
Not in favorable or adverse group poor initial response
FLT3 ITD+
Adverse −5, −7, 5q abn, 3q, 17p, 11q (other than 9;11), t(6;9), <50%; >90% Outcome worse in older patients, with t-AML, and
complex, monosomal karyotype, wild type NMP1 and poor initial response
FLT3-ITD, Mutated RUNX1, Mutated ASXL1 and
mutated TP53, insufficient metaphases for analysis
CR, Complete remission; HCT, hematopoietic cell transplant; NK, normal karyotype, +/+, double mutation.
and excluding MLL translocations (see later) (see Fig. 56.26). Because interrupts two genes—RUNX1 (CBFA2) on chromosome 21, band
of their specific association with distinct subtypes of leukemia, some q22, in intron 5, and RUNX1T1 (ETO [eight, twenty-one gene], on
AML-specific translocations have been incorporated into the WHO chromosome 8, band q22—and joins them to form a new chimeric
classification as criteria for subclassification of AML, including gene on the abnormal der(8) chromosome. RUNX1 is a gene on
t(8;21), t(15;17), inv(16), and 11q23 rearrangements, regardless of chromosome 21, also known as CBFA2 because it encodes for a
the morphology or percentage of blast cells. DNA-binding component of CBF and binds DNA through a specific
Two specific cytogenetic types of AML—t(8;21)(q22;q22) and sequence called the runt domain. The RUNX1 gene locus on chromo-
inv(16)(p13q22) or t(16;16)(p13;q22) (see Fig. 56.27)—are called some 21 spans 120 kb; the RUNX1T1 gene on chromosome 8 is
core-binding factor (CBF) AMLs and are usually grouped in clinical distributed over 87 kb.
studies because of similarities between their molecular and prognostic The RUNX1 gene has been identified in more than 39 chromo-
features. However, the morphologic features of these two AML somal translocations in leukemia and plays a critical role during
subtypes are very different, suggesting that individual approaches may hematopoiesis (see different RUNX1 chromosomal partners: http://
enhance clinical benefits in future studies. Patients with t(8;21) atlasgeneticsoncology.org/Genes/AML1ID52.html). The fusion gene is
exhibit large myeloid blasts with abundant basophilic cytoplasm, located on the partner chromosome in the majority of RUNX1
numerous azurophilic granules, and occasional Auer rods. Patients translocations. Its disruption is associated with the development of
with 16q22 abnormalities show variable numbers of eosinophils, myeloid and lymphoid leukemias. Therefore RUNX1 is viewed as a
usually in increased numbers, in all stages of maturation. The imma- master regulatory switch that controls development of a definitive
ture eosinophilic granules are larger than normal and may contain hematopoietic lineage. Moreover, the RUNX1 transcription factor is
purple-violet cytoplasmic granules. The t(8;21) subtype may be critical for proliferation and differentiation of hematopoietic stem
present alone, although 30% to 35% of patients also display loss of cells. Haploinsufficiency of RUNX1 has been linked to a propensity
Y chromosomes in males and loss of X chromosome in females (see to develop AML, and biallelic nonsense mutations in the RUNX1
Fig. 56.26, top left). Another 20% of patients with t(8;21) have a gene have been identified in the primitive AMLs of the French-
deletion of 9q12–23, including a commonly deleted segment that American-British (FAB) M0 subtype. The RUNX1T1 protein
spans 7 to 8 Mb. Trisomies for chromosomes 4 and 8 together with belongs to the ETO family of proteins involved in protein–protein
t(8;21) are observed in 6% to 10% of patients. Virtually all patients interactions but not in protein–DNA interactions.
with t(8;21) achieve CR. Additional cytogenetic abnormalities, irre- In the hybrid RUNX1-RUNXT1 protein, the C-terminus of
spective of their nature or complexity, do not have a deleterious effect RUNX1 is replaced by the entire RUNXT1 protein. The main
on rates of remission, relative risk of relapse, and OS. The t(8;21) functional characteristic of RUNX1-RUNX1T1 chimeric protein is