Page 920 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 920
Chapter 56 Conventional and Molecular Cytogenomic Basis of Hematologic Malignancies 803
t(16;16)(p13;q22)
8 der(21) 21 der(21) ider(21)
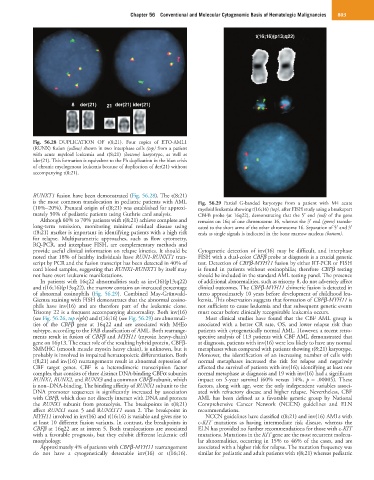
Fig. 56.28 DUPLICATION OF t(8;21). Four copies of ETO-AML1
(RUNX) fusion (yellow) shown in two interphase cells (top) from a patient
with acute myeloid leukemia and t(8;21) (bottom) karyotype, as well as
ider(21). This formation is equivalent to the Ph duplication in the blast crisis
of chronic myelogenous leukemia because of duplication of der(21) without
accompanying t(8;21).
RUNXT1 fusion have been demonstrated (Fig. 56.28). The t(8;21)
is the most common translocation in pediatric patients with AML Fig. 56.29 Partial G-banded karyotype from a patient with M4 acute
(10%–20%). Prenatal origin of t(8;21) was established for approxi- myeloid leukemia showing t(16;16) (top), after FISH study using a breakapart
mately 50% of pediatric patients using Guthrie card analysis. CBFB probe (at 16q22), demonstrating that the 5′ end (red) of the gene
Although 60% to 70% patients with t(8;21) achieve complete and remains on 16q of one chromosome 16, whereas the 3′ end (green) translo-
long-term remission, monitoring minimal residual disease using cated to the short arms of the other chromosome 16. Separation of 5′ and 3′
t(8;21) marker is important in identifying patients with a high risk ends as single signals is indicated in the bone marrow nucleus (bottom).
for relapse. Multiparametric approaches, such as flow cytometry,
RQ-PCR, and interphase FISH, are complementary methods and
provide useful clinical information on relapse kinetics. It should be Cytogenetic detection of inv(16) may be difficult, and interphase
noted that 18% of healthy individuals have RUN1-RUNXT1 tran- FISH with a dual-color CBFβ probe at diagnosis is a crucial genetic
script by PCR and the fusion transcript has been detected in 40% of test. Detection of CBFβ-MYH11 fusion by either RT-PCR or FISH
cord blood samples, suggesting that RUNX1-RUNXT1 by itself may is found in patients without eosinophilia; therefore CBFβ testing
not have overt leukemic manifestations. should be included in the standard AML testing panel. The presence
In patients with 16q22 abnormalities such as inv(16)(p13;q22) of additional abnormalities, such as trisomy 8, do not adversely affect
and t(16;16)(p13;q22), the marrow contains an increased percentage clinical outcomes. The CBFβ-MYH11 chimeric fusion is detected in
of abnormal eosinophils (Fig. 56.29). Combined May-Grünwald- utero approximately 10 years before development of childhood leu-
Giemsa staining with FISH demonstrates that the abnormal eosino- kemia. This observation suggests that formation of CBFβ-MYH11 is
phils have inv(16) and are therefore part of the leukemic clone. not sufficient to cause leukemia and that subsequent genetic events
Trisomy 22 is a frequent accompanying abnormality. Both inv(16) must occur before clinically recognizable leukemia occurs.
(see Fig. 56.26, top right) and t(16;16) (see Fig. 56.29) are abnormali- Most clinical studies have found that the CBF AML group is
ties of the CBFβ gene at 16q22 and are associated with M4Eo associated with a better CR rate, OS, and lower relapse risk than
subtype, according to the FAB classification of AML. Both rearrange- patients with cytogenetically normal AML. However, a recent retro-
ments result in fusion of CBFβ and MYH11 (myosin heavy-chain) spective analysis of 113 patients with CBF AML demonstrated that
gene on 16p13. The exact role of the resulting hybrid protein, CBFβ- at diagnosis, patients with inv(16) were less likely to have any normal
SMMHC (smooth muscle myosin heavy chain), is unknown, but it metaphases when compared with patients showing t(8;21) karyotype.
probably is involved in impaired hematopoietic differentiation. Both Moreover, the identification of an increasing number of cells with
t(8;21) and inv(16) rearrangements result in abnormal repression of normal metaphases increased the risk for relapse and negatively
CBF target genes. CBF is a heterodimeric transcription factor affected the survival of patients with inv(16); identifying at least one
complex that consists of three distinct DNA-binding CBFα subunits normal metaphase at diagnosis and 19 with inv(16) had a significant
RUNX1, RUNX2, and RUNX3 and a common CBFβ subunit, which impact on 5-year survival (60% versus 14%, p = .00005). These
is non–DNA-binding. The binding affinity of RUNX1 subunit to the factors, along with age, were the only independent variables associ-
DNA promoter sequences is significantly increased by association ated with refractory disease and higher relapse. Nevertheless, CBF
with CBFβ, which does not directly interact with DNA and protects AML has been defined as a favorable genetic group by National
the RUNX1 subunit from proteolysis. The breakpoints in t(8;21) Comprehensive Cancer Network (NCCN) guidelines and ELN
affect RUNX1 exon 5 and RUNX1T1 exon 2. The breakpoint in recommendations.
MYH11 involved in inv(16) and t(16;16) is variable and gives rise to NCCN guidelines have classified t(8;21) and inv(16) AMLs with
at least 10 different fusion variants. In contrast, the breakpoints in c-KIT mutations as having intermediate risk disease, whereas the
CBFβ at 16q22 are at intron 5. Both translocations are associated ELN has provided no further recommendations for those with c-KIT
with a favorable prognosis, but they exhibit different leukemic cell mutations. Mutations in the KIT gene are the most recurrent molecu-
morphology. lar abnormalities, occurring in 15% to 46% of the cases, and are
Approximately 4% of patients with CBFβ-MYH11 rearrangement associated with a higher risk for relapse. The mutation frequency was
do not have a cytogenetically detectable inv(16) or t(16;16). similar for pediatric and adult patients with t(8;21) whereas pediatric