Page 934 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 934
Chapter 56 Conventional and Molecular Cytogenomic Basis of Hematologic Malignancies 817
Low risk
A B C D
High risk
E
Very high risk
F G H
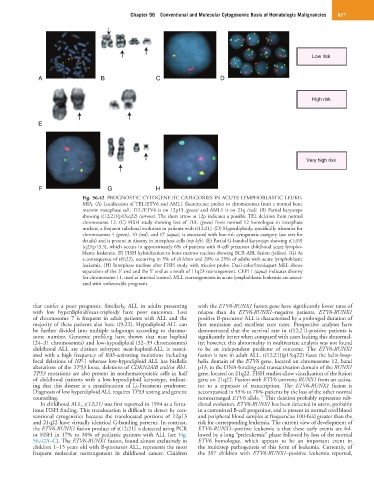
Fig. 56.42 PROGNOSTIC CYTOGENETIC CATEGORIES IN ACUTE LYMPHOBLASTIC LEUKE-
MIA. (A) Localization of TEL/ETV6 and AML1 fluorescence probes to chromosomes from a normal bone
marrow metaphase cell. TEL/ETV6 is on 12p13 (green) and AML1 is on 21q (red). (B) Partial karyotype
showing t(12;21)(p13;q22) (arrows). The short arrow at 12p indicates a possible TEL deletion from normal
chromosome 12. (C) FISH study showing loss of TEL (green) from normal 12 homologue in interphase
nucleus, a frequent subclonal evolution in patients with t(12;21). (D) Hyperdiploidy, specifically trisomies for
chromosomes 4 (green), 10 (red), and 17 (aqua), is associated with low-risk cytogenetic category (see text for
details) and is present in disomy in interphase cells (top left). (E) Partial G-banded karyotype showing t(1;19)
(q23;p13.3), which occurs in approximately 6% of patients with B-cell precursor childhood acute lympho-
blastic leukemia. (F) FISH hybridization to bone marrow nucleus showing BCR-ABL fusion (yellow). (G) As
a consequence of t(9;22), occurring in 5% of children and 20% to 25% of adults with acute lymphoblastic
leukemia. (H) Interphase nucleus after FISH study with tricolor probe. Dual-color/breakapart MLL shows
separation of the 3′ end and the 5′ end as a result of 11q23 rearrangement. CEP11 (aqua) indicates disomy
for chromosome 11, used as internal control. MLL rearrangements in acute lymphoblastic leukemia are associ-
ated with unfavorable prognosis.
that confer a poor prognosis. Similarly, ALL in adults presenting with the ETV6-RUNX1 fusion gene have significantly lower rates of
with low hyperdiploid/near-triploidy have poor outcomes. Loss relapse than do ETV6-RUNX1–negative patients. ETV6-RUNX1
of chromosome 7 is frequent in adult patients with ALL and the positive B-precursor ALL is characterized by a prolonged duration of
majority of these patients also have t(9;22). Hypodiploid ALL can first remission and excellent cure rates. Prospective analyses have
be further divided into multiple subgroups according to chromo- demonstrated that the survival rate in t(12;21)-positive patients is
some number. Genomic profiling have shown that near haploid significantly better when compared with cases lacking this abnormal-
(24–31 chromosomes) and low-hypodiploid (32–39 chromosomes) ity; however, this abnormality in multivariate analysis was not found
childhood ALL are distinct subtypes: near-haploid-ALL is associ- to be an independent predictor of outcome. The ETV6-RUNX1
ated with a high frequency of RAS-activating mutations including fusion is rare in adult ALL. t(12;21)(p13;q22) fuses the helix-loop-
focal deletions of NF-1 whereas low-hypodiploid ALL has biallelic helix domain of the ETV6 gene, located on chromosome 12, band
alterations of the TP53 locus, deletions of CDKN2A/B and/or Rb1. p13, to the DNA-binding and transactivation domain of the RUNX1
TP53 mutations are also present in nonhematopoietic cells in half gene, located on 21q22. FISH studies allow visualization of the fusion
of childhood patients with a low-hyperdiploid karyotype, indicat- gene on 21q22. Fusion with ETV6 converts RUNX1 from an activa-
ing that this disease is a manifestation of Li-Fraumeni syndrome. tor to a repressor of transcription. The ETV6-RUNX1 fusion is
Diagnosis of low hyperdiploid ALL requires TP53 testing and genetic accompanied in 55% to 70% patients by the loss of the other normal
17
counseling. nonrearranged ETV6 allele. This deletion probably represents sub-
In childhood ALL, t(12;21) was first reported in 1994 as a fortu- clonal evolution. ETV6-RUNX1 has been detected in utero, probably
itous FISH finding. This translocation is difficult to detect by con- in a committed B-cell progenitor, and is present in normal cord blood
ventional cytogenetics because the translocated portions of 12p13 and peripheral blood samples at frequencies 100-fold greater than the
and 21q22 have virtually identical G-banding patterns. In contrast, risk for corresponding leukemia. The current view of development of
the ETV6-RUNX1 fusion product of t(12;21) is detected using PCR ETV6-RUNX1–positive leukemia is that these early events are fol-
or FISH in 17% to 30% of pediatric patients with ALL (see Fig. lowed by a long “preleukemic” phase followed by loss of the normal
56.42A–C). The ETV6-RUNX1 fusion, found almost exclusively in ETV6 homologue, which appears to be an important event in
children 1–15 years old with B-precursor ALL, represents the most the multistep pathogenesis of this form of leukemia. Currently, of
frequent molecular rearrangement in childhood cancer. Children the 397 children with ETV6-RUNX1–positive leukemia reported,