Page 936 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 936
Chapter 56 Conventional and Molecular Cytogenomic Basis of Hematologic Malignancies 819
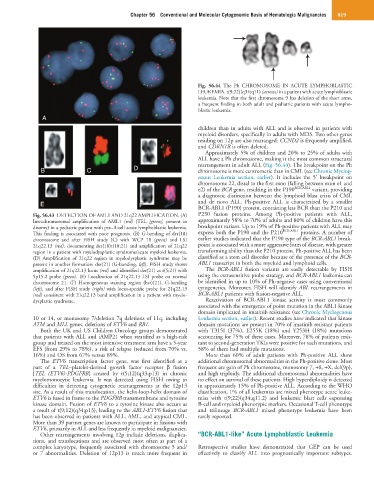
Fig. 56.44 The Ph CHROMOSOME IN ACUTE LYMPHOBLASTIC
LEUKEMIA. t(9;22)(q34;q11) (arrows) in a patient with acute lymphoblastic
leukemia. Note that the first chromosome 9 has deletion of the short arms,
a frequent finding in both adult and pediatric patients with acute lympho-
blastic leukemia.
A
children than in adults with ALL and is observed in patients with
myeloid disorders, specifically in adults with MDS. Two other genes
residing on 12p are also rearranged: CCND2 is frequently amplified,
and CDKN1B is often deleted.
Approximately 5% of children and 20% to 25% of adults with
ALL have a Ph chromosome, making it the most common structural
B C D rearrangement in adult ALL (Fig. 56.44). The breakpoint on the Ph
chromosome is more centromeric than in CML (see Chronic Myelog-
enous Leukemia section, earlier). It includes the 5′ breakpoint on
chromosome 22, distal to the first exon (falling between exon e1 and
e2) of the BCR gene, resulting in the P190 BCR-ABL1 variant, providing
a diagnostic distinction between the lymphoid blast crisis of CML
E F F and de novo ALL. Ph-positive ALL is characterized by a smaller
BCR-ABL1 (P190) protein, containing less BCR than the P210 and
Fig. 56.43 DETECTION OF AML1 AND 21q22 AMPLIFICATION. (A) P230 fusion proteins. Among Ph-positive patients with ALL,
Intrachromosomal amplification of AML1 (red) (TEL [green], present in approximately 58% to 70% of adults and 80% of children have this
disomy) in a pediatric patient with pre–B-cell acute lymphoblastic leukemia. breakpoint variant. Up to 19% of Ph-positive patients with ALL may
BCR-ABL1
This finding is associated with poor prognosis. (B) G-banding of der(18) express both the P190 and the P210 proteins. A number of
chromosome and after FISH study (C) with WCP 18 (green) and LSI earlier studies indicated that the P190 type of the BCR-ABL1 break-
21q22.13 (red), documenting der(18)t(18;21) and amplification of 21q22 point is associated with a more aggressive form of disease, with greater
region in a patient with myelodysplastic syndrome/acute myeloid leukemia. transforming ability than the P210 protein. Ph-positive ALL has been
(D) Amplification of 21q22 region in myelodysplastic syndrome may be classified as a stem cell disorder because of the presence of the BCR-
present in another formation: der(21) (G-banding, left). FISH study shows ABL1 transcript in both the myeloid and lymphoid cells.
amplification of 21q22.13 locus (red) and identified der(21) as t(5;21) with The BCR-ABL1 fusion variants are easily detectable by FISH
5p15.2 probe (green). (E) Localization of 21q22.13 LSI probe on normal using the extrasensitive probe strategy, and BCR-ABL1 leukemia can
chromosome 21. (F) Homogeneous staining region (hsr)(21), G-banding be identified in up to 10% of Ph-negative cases using conventional
(left), and after FISH study (right) with locus-specific probe for 21q22.13 cytogenetics. Moreover, FISH will identify ABL rearrangements in
(red) consistent with 21q22.13 band amplification in a patient with myelo- BCR-ABL1 patients with fusion-negative ALL.
dysplastic syndrome. Reactivation of BCR-ABL1 kinase activity is most commonly
associated with the emergence of point mutation in the ABL1 kinase
domain implicated in imatinib resistance (see Chronic Myelogenous
10 or 14, or monosomy 7/deletion 7q deletions of 11q, including Leukemia section, earlier). Recent studies have indicated that kinase
ATM and MLL genes, deletions of ETV6 and RB1. domain mutations are present in 70% of imatinib resistant patients
Both the UK and US Children Oncology groups demonstrated with T315I (37%), E255K (18%) and Y253H (18%) mutations
that patients with ALL and iAMP21 when stratified as a high-risk accounting for 75% of these cases. Moreover, 78% of patients resis-
group and treated on the most intensive treatment arm have a 5-year tant to second-generation TKIs were positive for such mutations, and
EFS (from 29% to 78%), a risk of relapse (reduced from 70% vs. 58% of them had multiple mutations.
16%) and OS from 67% versus 89%. More than 60% of adult patients with Ph-positive ALL show
The ETV6 transcription factor gene, was first identified as a additional chromosomal abnormalities in the Ph-positive clone. Most
part of a TEL–platelet-derived growth factor receptor β fusion frequent are gain of Ph chromosome, monosomy 7, +8, +X, del(9p),
[TEL (ETV6)-PDGFRB] created by t(5;12)(q33;p13) in chronic and high triploidy. The additional chromosomal abnormalities have
myelomonocytic leukemia. It was detected using FISH owing to no effect on survival of these patients. High hyperdiploidy is detected
difficulties in detecting cytogenetic rearrangements at the 12p13 in approximately 15% of Ph-positive ALL. According to the WHO
site. As a result of this translocation, the helix-loop-helix domain of classification, 1% of all leukemias are mixed phenotype acute leuke-
ETV6 is fused in frame to the PDGFRB transmembrane and tyrosine mias with t(9;22)(q34;q11.2) and leukemic blast cells expressing
kinase domain. Fusion of ETV6 to a tyrosine kinase also occurs as B-cell and myeloid phenotypic markers. Occasional T-cell phenotype
a result of t(9;12)(q34;p13), leading to the ABL1-ETV6 fusion that and trilineage BCR-ABL1 mixed phenotype leukemia have been
has been observed in patients with ALL, AML, and atypical CML. rarely reported.
More than 39 partner genes are known to participate in fusions with
ETV6, primarily in ALL and less frequently in myeloid malignancies.
Other rearrangements involving 12p include deletions, duplica- “BCR-ABL1-like” Acute Lymphoblastic Leukemia
tions, and translocations and are observed most often as part of a
complex karyotype, frequently associated with chromosome 5 and/ Retrospective studies have demonstrated that GEP can be used
or 7 abnormalities. Deletion of 12p13 is much more frequent in effectively to classify ALL into prognostically important subtypes.