Page 941 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 941
824 Part VII Hematologic Malignancies
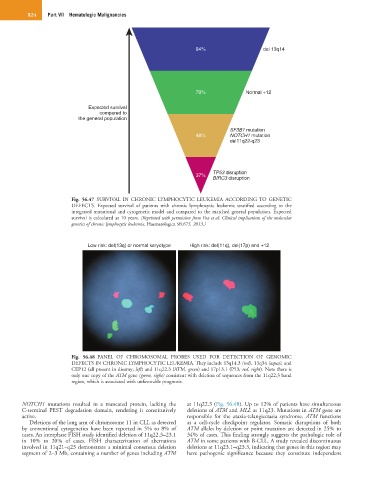
84% del 13q14
70% Normal +12
Expected survival
compared to
the general population
SF3B1 mutation
48% NOTCH1 mutation
del11q22-q23
TP53 disruption
37%
BIRC3 disruption
Fig. 56.47 SURVIVAL IN CHRONIC LYMPHOCYTIC LEUKEMIA ACCORDING TO GENETIC
DEFECTS. Expected survival of patients with chronic lymphocytic leukemia stratified according to the
integrated mutational and cytogenetic model and compared to the matched general population. Expected
survival is calculated at 10 years. (Reprinted with permission from Foa et al: Clinical implications of the molecular
genetics of chronic lymphocytic leukemia. Haematologica 98:675, 2013.)
Low risk: del(13q) or normal karyotype High risk: del(11q), del(17p) and +12
Fig. 56.48 PANEL OF CHROMOSOMAL PROBES USED FOR DETECTION OF GENOMIC
DEFECTS IN CHRONIC LYMPHOCYTIC LEUKEMIA. They include 13q14.3 (red), 13q34 (aqua), and
CEP12 (all present in disomy; left) and 11q22.3 (ATM, green) and 17p13.1 (P53; red, right). Note there is
only one copy of the ATM gene (green, right) consistent with deletion of sequences from the 11q22.3 band
region, which is associated with unfavorable prognosis.
NOTCH1 mutations resulted in a truncated protein, lacking the at 11q22.3 (Fig. 56.48). Up to 12% of patients have simultaneous
C-terminal PEST degradation domain, rendering it constitutively deletions of ATM and MLL at 11q23. Mutations in ATM gene are
active. responsible for the ataxia-telangiectasia syndrome. ATM functions
Deletions of the long arm of chromosome 11 in CLL as detected as a cell-cycle checkpoint regulator. Somatic disruptions of both
by conventional cytogenetics have been reported in 5% to 8% of ATM alleles by deletion or point mutation are detected in 25% to
cases. An interphase FISH study identified deletion of 11q22.3–23.1 34% of cases. This finding strongly suggests the pathologic role of
in 10% to 20% of cases. FISH characterization of aberrations ATM in some patients with B-CLL. A study revealed discontinuous
involved in 11q21–q23 demonstrate a minimal consensus deletion deletions at 11q23.1–q23.3, indicating that genes in this region may
segment of 2–3 Mb, containing a number of genes including ATM have pathogenic significance because they constitute independent