Page 940 - Hematology_ Basic Principles and Practice ( PDFDrive )
P. 940
Chapter 56 Conventional and Molecular Cytogenomic Basis of Hematologic Malignancies 823
TABLE Most Frequent Clinically Relevant Chromosomal Abnormalities and Copy Number Alterations in Chronic Lymphocytic Leukemia
56.11
Chromosomal Clinical Associations and
Abnormality Frequency % Likely Gene Target Consequence miRNAs Prognosis
amp(2p) 7 REL, XPO1, BCL11A Unknown Poor
amp(3q26.32) 6 PIK3CA Unknown Poor
del(6q) ~6 Unknown Associated with prominent Poor
lymphocytosis
Atypical morphology
Splenomegaly
Higher rates of CD38 positivity
del(8p) 5 Unknown Unknown Poor
amp(8)(q24.21) 5 MYC Poor
del(10)(q24) 2 NFkB2 Unknown Unknown
del(11)(q22.3) 10–20 ATM, BIRC3 Defect in DNA repair ↑miR-29b Poor
Deregulation of P53 ↑miR-155
Deregulation of cell cycle miR-29a
miR-34b
Extensive lymphadenopathy miR-34c
Trisomy 12 10–23 Unknown Atypical morphology ↑miR-148a Poor
Aggressive clinical phenotype ↑miR-146a
del(13)(q14) 57–61 miR15a/16 in an intron of DLEU2 BCL2 expression ↑miR-155 Good
Its deletion leads to the release of BCL2 ↑miR-7–1
Resistance to apoptosis ↑miR-154
↓miR-220
↓miR-221
del(15)(q15.1) 4 MGA Unknown None
del(17)(p13.1) 6–8 TP53 Defect in DNA repair ↑miR-151 Poor
Deregulation of cell cycle ↓miR29C
Aggressive clinical phenotype ↓miR34a
↓miR148a
↓miR-181
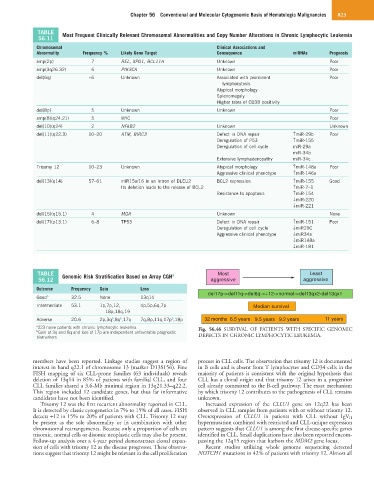
TABLE Genomic Risk Stratification Based on Array CGH a Most Least
56.12 aggressive aggressive
Outcome Frequency Gain Loss
del17p->del11q->del6q->+12->normal->del13qx2-del13qx1
Good b 32.5 None 13q14
Intermediate 53.1 1p,7p,12, 4p,5p,6q,7p Median survival
18p,18q,19
b
b
Adverse 20.6 2p,3q ,8q ,17q 7q,8p,11q,17p ,18p 32 months 6.5 years 9.5 years 9.2 years 11 years
b
a 223 naive patients with chronic lymphocytic leukemia. Fig. 56.46 SURVIVAL OF PATIENTS WITH SPECIFIC GENOMIC
b Gain of 3q and 8q and loss of 17p are independent unfavorable prognostic
biomarkers. DEFECTS IN CHRONIC LYMPHOCYTIC LEUKEMIA.
members have been reported. Linkage studies suggest a region of process in CLL cells. The observation that trisomy 12 is documented
interest in band q22.1 of chromosome 13 (marker D13S156). Fine in B cells and is absent from T lymphocytes and CD34 cells in the
FISH mapping of six CLL-prone families (63 individuals) reveals majority of patients is consistent with the original hypothesis that
deletion of 13q14 in 85% of patients with familial CLL, and four CLL has a clonal origin and that trisomy 12 arises in a progenitor
CLL families shared a 3.6-Mb minimal region in 13q21.33–q22.2. cell already committed to the B-cell pathway. The exact mechanism
This region included 12 candidate genes, but thus far informative by which trisomy 12 contributes to the pathogenesis of CLL remains
candidates have not been identified. unknown.
Trisomy 12 was the first recurrent abnormality reported in CLL. Increased expression of the CLLU1 gene on 12q22 has been
It is detected by classic cytogenetics in 7% to 15% of all cases. FISH observed in CLL samples from patients with or without trisomy 12.
detects +12 in 15% to 20% of patients with CLL. Trisomy 12 may Overexpression of CLLU1 in patients with CLL without IgV H
be present as the sole abnormality or in combination with other hypermutation combined with restricted and CLL-unique expression
chromosomal rearrangements. Because only a proportion of cells are pattern suggests that CLLU1 is among the first disease-specific genes
trisomic, normal cells or disomic neoplastic cells may also be present. identified in CLL. Small duplications have also been reported encom-
Follow-up analysis over a 4-year period demonstrates clonal expan- passing the 12q15 region that harbors the MDM2 gene locus.
sion of cells with trisomy 12 as the disease progresses. These observa- Recent studies utilizing whole genome sequencing detected
tions suggest that trisomy 12 might be relevant in the cell proliferation NOTCH1 mutations in 42% of patients with trisomy 12. Almost all