Page 325 - Williams Hematology ( PDFDrive )
P. 325
300 Part IV: Molecular and Cellular Hematology Chapter 20: Innate Immunity 301
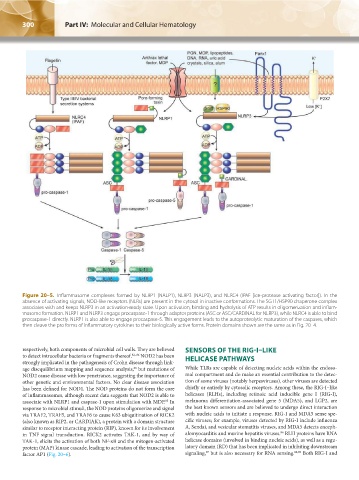
Figure 20–5. Inflammasome complexes formed by NLRP1 (NALP1), NLRP3 (NALP3), and NLRC4 (IPAF [ice-protease activating factor]). In the
absence of activating signals, NOD-like receptors (NLRs) are present in the cytosol in inactive conformations. The SGT1/HSP90 chaperone complex
associates with and keeps NLRP3 in an activation-ready state. Upon activation, binding and hydrolysis of ATP results in oligomerization and inflam-
masome formation. NLRP1 and NLRP3 engage procaspase-1 through adaptor proteins (ASC or ASC/CARDINAL for NLRP3), while NLRC4 is able to bind
procaspase-1 directly. NLRP1 is also able to engage procaspase-5. This engagement leads to the autoproteolytic maturation of the caspases, which
then cleave the pro forms of inflammatory cytokines to their biologically active forms. Protein domains shown are the same as in Fig. 20–4.
respectively, both components of microbial cell walls. They are believed SENSORS OF THE RIG-I–LIKE
to detect intracellular bacteria or fragments thereof. 82–84 NOD2 has been HELICASE PATHWAYS
strongly implicated in the pathogenesis of Crohn disease through link-
age disequilibrium mapping and sequence analysis, but mutations of While TLRs are capable of detecting nucleic acids within the endoso-
66
NOD2 cause disease with low penetrance, suggesting the importance of mal compartment and do make an essential contribution to the detec-
other genetic and environmental factors. No clear disease association tion of some viruses (notably herpesviruses), other viruses are detected
has been defined for NOD1. The NOD proteins do not form the core chiefly or entirely by cytosolic receptors. Among these, the RIG-I–like
of inflammasomes, although recent data suggests that NOD2 is able to helicases (RLHs), including retinoic acid inducible gene I (RIG-I),
associate with NLRP1 and caspase-1 upon stimulation with MDP. In melanoma differentiation-associated gene 5 (MDA5), and LGP2, are
85
response to microbial stimuli, the NOD proteins oligomerize and signal the best known sensors and are believed to undergo direct interaction
via TRAF2, TRAF5, and TRAF6 to cause K63 ubiquitination of RICK2 with nucleic acids to initiate a response. RIG-I and MDA5 sense spe-
(also known as RIP2, or CARDIAK), a protein with a domain structure cific viruses; for example, viruses detected by RIG-I include influenza
similar to receptor interacting protein (RIP), known for its involvement A, Sendai, and vesicular stomatitis viruses, and MDA5 detects enceph-
86
in TNF signal transduction. RICK2 activates TAK-1, and by way of alomyocarditis and murine hepatitis viruses. RLH proteins have RNA
TAK-1, elicits the activation of both NF-κB and the mitogen-activated helicase domains (involved in binding nucleic acids), as well as a regu-
protein (MAP) kinase cascade, leading to activation of the transcription latory domain (RD) that has been implicated in inhibiting downstream
87
factor AP1 (Fig. 20–6). signaling, but is also necessary for RNA sensing. 88,89 Both RIG-I and
Kaushansky_chapter 20_p0293-0306.indd 300 9/17/15 5:52 PM